Abstract
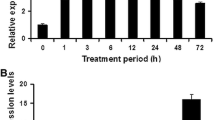
In this report, we present data on OsSDS1 (Oryza sativa L. salt and drought sensitive gene 1)—an uncharacterized gene isolated from rice Pei’ai 64S (O. sativa L.). Expression of OsSDS1 was strongly up-regulated by a wide spectrum of stresses, including cold, drought, and heat, in different tissues at different developmental stages of rice, as revealed by both microarray and quantitative RT-PCR analyses. Subcellular localization revealed that an OsSDS1: GFP fusion protein was distributed to the nucleus. Expression of OsSDS1 conferred decreased tolerance to salt and drought in Arabidopsis thaliana, accompanied by altered expression of stress-responsive genes and altered K+/Na+ ratio. The results suggest that OsSDS1 may act as a negative regulator of salt and drought tolerance in plants.






Similar content being viewed by others
Abbreviations
- CaMV:
-
Cauliflower mosaic virus
- GFP:
-
Green fluorescent protein
- Pn:
-
Net photosynthesis rate
- qRT-PCR:
-
Quantitative real-time RT-PCR
- WT:
-
Wild type
References
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311:91–94
Apse MP, Blumwald E (2002) Engineering salt tolerance in plants. Curr Opin Biotechnol 13:146–150
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285(5431):1256–1258
Bi CL, Wen XJ, Zhang XY, Liu X (2010) Cloning and characterization of a putative CTR1 gene from wheat. Agric Sci China 9(9):1241–1250
Chinnusamy V, Zhu J, Zhu JK (2006) Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng (N Y) 27:141–177
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Hadiarto T, Tran LS (2011) Progress studies of drought-responsive genes in rice. Plant Cell Rep 30:297–310
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Jogaiah S, Govind SR, Tran LS (2012) System biology-based approaches towards understanding drought tolerance in food crops. Crit Rev Biotechnol. doi:10.3109/07388551.2012.659174
Lu GJ, Gao CX, Zheng XN, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229(3):605–615
Luo LJ (2010) Breeding for water-saving and drought-resistance rice (WDR) in China. J Exp Bot 61:3509–3517
Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M (2010) The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J 63:484–498
Muñoz-Mayor A, Pineda B, Garcia-Abellán JO, Antón T, Garcia-Sogo B, Sanchez-Bel P, Flores FB, Atarés A, Angosto T, Pintor-Toro JA, Moreno V, Bolarin MC (2012) Overexpression of dehydrin tas14 gene improves the osmotic stress imposed by drought and salinity in tomato. J Plant Physiol 169:459–468
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco cultures. Physiol Plant 15:473–497
Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K (2011) SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol 157:1900–1913
Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol Plant 1:538–551
Qu GZ, Zang LN, Hu XL, Gao CQ, Zheng TC, Li KL (2012) Co-transfer of LEA and bZip genes from tamarix confers additive salt and osmotic stress tolerancein transgenic tobacco. Plant Mol Biol Rep 30:512–518
Ren Z, Zheng Z, Chinnusamy V, Zhu J, Cui X, Iida K, Zhu JK (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 107:5669–5674
Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14:465–477
Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T (2005) LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol 46:1623–1634
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Tao Z, Kou YJ, Liu HB, Li XH, Xiao JH, Wang SP (2011) OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62:4863–4874
Varagona MJ, Schmidt RJ, Raikhel NV (1992) Nuclear localization signals required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4:1213–1227
Varet A, Parker J, Tornero P, Nass N, Nürnberger T, Dangl JL, Scheel D, Lee J (2002) NHL25 and NHL3, two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol Plant Microbe In 15:608–616
Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic Leucine Zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148:1938–1952
Xu K, Hong P, Luo LJ, Xia T (2010) Overexpression of AtNHX1, a vacuolar Na+/H+ antiporter from Arabidopsis thaliana, in Petunia hybrida enhances salt and drought tolerance. J Plant Biol 52:453–461
Xu GY, Rocha P, Wang ML, Xu ML, Cui YC, Li LY, Zhu YX, Xia XJ (2011) A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 234:47–59
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yoshiba Y, Nanjo T, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Stress-responsive and developmental regulation of Delta(1)-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem Biophys Res Commun 261:766–772
Zhang X, Guo XP, Lei CL, Cheng ZJ, Lin QB, Wang JL, Wu FQ, Wang J, Wan JM (2011a) Overexpression of SlCZFP1, a novel TFIIIA-type Zinc Finger protein from tomato, confers enhanced cold tolerance in transgenic Arabidopsis and rice. Plant Mol Biol Rep 29:185–196
Zhang CL, Shi HJ, Chen L, Wang XM, Lu BB, Zhang SP, Liang Y, Liu RX, Qian J, Sun WW, You ZZ, Dong HS (2011b) Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol 11:11
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Acknowledgments
This research was supported by Nitrogen and Phosphorus cycling and manipulation for agro-ecosystems and the Knowledge Innovation program of the Chinese Academy of Sciences (KZCX2-YW-T07) and National Natural Science Foundation of China (31171536).
Author information
Authors and Affiliations
Corresponding author
Additional information
G. Xu and M. Li contributed equally to this work
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 65 kb)
Rights and permissions
About this article
Cite this article
Xu, G., Li, M., Huang, M. et al. The Negative Regulator OsSDS1 Significantly Reduces Salt and Drought Tolerance in Transgenic Arabidopsis. Plant Mol Biol Rep 31, 517–523 (2013). https://doi.org/10.1007/s11105-012-0521-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0521-8