Abstract
Cellulose is the major component of plant cell walls, providing mechanical strength to the structural framework of plants. In association with lignin, hemicellulose, protein and pectin, cellulose forms the strong yet flexible bio-composite tissue of wood. Wood formation is an essential biological process and is of significant importance to the cellulosic private sector industry. Cellulose synthase genes encode the catalytic subunits of a large protein complex responsible for the biogenesis of cellulose in higher plants. The hybrid Acacia auriculiformis x Acacia mangium represents an important source of tree cellulose for forest-based product manufacturing, with enormous economic potential. In this work, we isolate the first cellulose synthase gene, designated AaxmCesA1, from this species. The isolated full-length AaxmCesA1 cDNA encodes a polypeptide of 1,064 amino acids. Sequence analyses revealed that AaxmCesA1 cDNA possesses the key motif characteristics of a CesA protein. AaxmCesA1 shares more than 75 % amino acid sequence identity with CesA proteins from other plant species. Subsequently, the full-length AaxmCesA1 gene of 7,389 bp with partial regulatory and 13 intron regions was also isolated. Relative gene expression analysis by quantitative PCR in different tissues of the Acacia hybrid, suggests the involvement of the AaxmCesA1 gene in primary cell wall synthesis of rapidly dividing young root cells. Similarity analyses using Blast algorithms also suggests a role in primary cell wall deposition in the Acacia hybrid. Southern analysis predicts that AaxmCesA1 is a member of a multigene family with at least two isoforms in the genome of the Acacia hybrid.
Similar content being viewed by others
Introduction
Acacia is a genus of shrubs and trees consisting of approximately 1,400 species found worldwide, with more than 900 species native to Australia. Acacia species spread around the tropical-warm–temperate regions of both hemispheres, including Southern Asia, Africa and the Americas (Maslin et al. 2003). Many Australia tropical acacias have substantial commercial importance for the pulp and timber industries, particularly in Southeast Asia. The main Acacia species with high commercial values are Acacia mangium, A. auriculiformis, A. crassicarpa and the more recently emerging A. auriculiformis x A. mangium hybrid (McDonald et al. 2001). In Malaysia, the first natural hybridization of Acacia auriculiformis x A. mangium hybrid (hereafter Acacia hybrid) was observed in an Acacia plantation in Ulu Kukut, Sabah in 1971 (Tham 1976). Better stem straightness compared to A. mangium was found in the hybrid; the hybrid also inherits better self-pruning ability, stem roundness and disease resistance than A. auriculiformis. The hybrid also possesses intermediate characteristics between the two parental Acacia species (Bowen 1981) and appears to have higher cellulose and lower lignin contents, which give better pulp yields than both parental trees (Yamada et al. 1990).
Cellulose is the major component in wood, which is present universally in plant cell walls and provides mechanical strength to the plant. Cellulose accounts for about 20 % and 50 % of the primary and secondary cell walls in higher plants, respectively. Membrane-bound cellulose synthase enzyme complexes (CelS) are believed to play a key role in the biosynthesis of cellulose (Saxena and Brown 2005). These complexes are discernible as hexameric rosettes under freeze-fracture electron microscopy and consist of 36 cellulose synthase (CesA) proteins per rosette (Doblin et al. 2002; Saxena and Brown 2005; Somerville 2006). Since the first isolation of a plant CesA gene from a cotton tree 16 years ago, many similar attempts have been made in numerous plants with the aim of understanding the role of CesA genes and the mechanisms involved during cellulose biosynthesis. Previous studies revealed that the catalytic subunits of CelS in many plant species are encoded by a family of genes or isoforms (Richmond and Somerville 2000; Djerbi et al. 2005). Isoforms are alternative forms of a gene generated through splicing (Wang et al. 2012) and are found commonly in many plant species. Isoforms with differential expression patterns in different types of tissues throughout different plant developmental stages have also been reported (Hayashi et al. 2005; Joshi et al. 2004; Nairn and Haselkorn 2005; Roberts et al. 2004; Taylor et al. 2003). Mutant analyses conducted on ten CesA genes from Arabidopsis thaliana (AtCesAs) also revealed a number of glycosyltransferases encoded by these AtCesAs that function in cellulose biosynthesis in primary and secondary cell walls (Burn et al. 2002; Taylor et al. 1999; Turner and Somerville 1997). A study on Eucalyptus had also identified several CesAs associated with primary and secondary cell wall formations (Ranik and Myburg 2006). Cellulose biogenesis is a complex process, which still requires ongoing study to comprehend completely its pathways and mechanisms. Even with the completion of the Arabidopsis thaliana genome sequencing project, researchers still struggle to understand the biogenesis of cellulose in A. thaliana, let alone other plants that are less well studied.
Despite being cultivated for its timber, as well as for the pulp and paper industries, not much is known about the genetics underlying cellulose biosynthesis in the Acacia hybrid. Here, we report the first isolation and characterization of a cellulose synthase gene from the commercially valuable Acacia hybrid with the aim of understanding the deposition of cellulose in its primary and secondary cell walls.
Materials and Methods
Plant Material and Total RNA Preparation
Young leaves, a mixture of phloem and xylem (hereafter referred to as inner bark), flowers and greenish seedpod tissues were collected from a 7-year-old Acacia hybrid tree (AaHyF1113) planted in Plot W, Plant Biotechnology Laboratory, Universiti Kebangsaan Malaysia. Young root tissue was obtained from tissue-cultured Acacia hybrid (Clone M5) provided by the Forest Research Institute Malaysia. Tissues were immersed in liquid nitrogen immediately upon collection. Total RNA was extracted from the five tissues using a RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations with minor modification, where 1 % polyethylene glycol 6000 (PEG 6000) was added to the protocol to enhance total RNA yield. Additional on-column DNase digestions were performed twice using RNase free DNase I (Promega, Madison, WI). The concentration and quality of the total RNA isolated was estimated from the ratio of the absorbance measured at 260/280 nm, and by gel electrophoresis. cDNA was synthesized using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA) and cDNA quality was assessed by end point reverse transcription PCR using gene specific primers (GSP).
Relative Gene Expression Analysis by Quantitative PCR
GSP for AaxmCesA1 and actin genes were designed based on the 3′ EST sequences identified from our Acacia hybrid EST dataset (Yong et al. 2011), using Primer Premier 5 (http://www.PremierBiosoft.com) and Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3) software, respectively. The primer sequences for CesA and actin genes were: CesAForw1: 5′ CATACCCGTTTGTCGTCC, CesARerv1: 5′ CATCAACATTCATCCTCATTG and ActinForw: 5′ GGTAACATTGTCCTCTGTGGT, ActinRerv: 5′ CATCGTATTCTGCCTTCGA. The relative expression level of AaxmCesA1 gene was investigated in the inner bark, young seedpod, leaf, young root and flower tissues, with the inner bark tissue as calibrator sample. PCR reactions were carried out using SYBR Green Supermix (Bio-Rad, Hercules, CA) in a iCycler iQ5 Real-Time PCR Detection System (Bio-Rad). PCR was initiated with a pre-denaturation step at 95 °C for 4 min, followed by 60 cycles of denaturation, annealing and extension at 95 °C/15 s, 55–60 °C/30s, 72 °C/15 s, respectively, then a final extension at 72 °C for 4 min. All standard dilutions, controls and samples were performed in four replicates. Standard curves for AaxmCesA1 and actin were constructed using 5-fold serial dilution of the RT product synthesized from inner bark tissue. The 10μL quantitative PCR (qPCR) reactions contained a final concentration of 1x SYBR Supermix, 0.2 μm of each primer and 10 ng template cDNA. Melt curve analysis was carried out from 55 °C to 95 °C at the end of the amplification to confirm that the individual qPCR product corresponded to a single homogeneous DNA species. qPCR products were also sequenced directly using ABI 3700 (Applied Biosystems, Foster City, CA) at Macrogen Corporation, Korea, to ensure amplification specificity.
Isolation of Full-Length AaxmCesA1 cDNA
Total RNA was extracted from the leaf samples of a single Acacia hybrid individual plant (HyAa/F1 113) using an RNeasy Plant Mini Kit (Qiagen). The full-length AaxmCesA1 cDNA was isolated using a BD SMART™ RACE cDNA amplification kit (Clontech, Palo Alto, CA) according to manufacturer’s instructions. RACE primers were designed using Primer Premier 5 software. The primers of RamA1: 5′ CCTTCCGAGCAGACCCTTCA, RCesA1-1Rb: 5′ GCCTGCTAAGGAAAATCTCAATGGACC and NRA11R: 5′ GCAATAGGCACCACAGACTCAAAGG were used for 5′ RACE PCR; FAMA1: 5′ GGTGGCATTCCTCCCTCAAC was used for 3′ RACE PCR. Each RACE-PCR reaction containing 2.5μL first strand cDNA, 1x UPM A Mix, 0.2 μM GSP and 41.5μL master mix was performed using the Mastercycler PCR machine (Eppendorf, Hauppauge, NY). PCR was performed using a touch down cycling profile starting with 5 cycles of amplification at 94 °C/30s, 72 °C/3 min, followed by another 5 cycles at 94 °C/30s, 70 °C/30s, 72 °C/3 min, then 25 cycles at 94 °C/30s, 68 °C/30s, 72 °C/4 min, and finally an extension at 72 °C for 16 min. RACE PCR products were electrophoresed on 1.2 % agarose gel in 1 x TAE buffer at 80 V, gel purified using Qiaquick Gel extraction kit (Qiagen) and cloned in the PGEM-T easy Vector system (Promega), before sequencing from both ends using ABI 3700 (Applied Biosystems) at Macrogen Corporation, Korea.
Isolation of AaxmCesA1 Intron Regions
Genomic DNA was extracted from the young leaf of an individual Acacia hybrid (AaHy113F1) using the CTAB method (Doyle and Doyle 1990). DNA sequences obtained from our RACE PCR were used to design primers (Table 1) using Primer Premier 5 and Primer 3 software for the amplification of the intron regions. Each 50μL PCR reaction contained 20–40 ng genomic DNA, 1x PCR buffer, 10 mM dNTP mix, 5uM of each primer and 1 U Hot Start Taq polymerase. PCR was performed in Mastercycler PCR machine (Eppendorf) using a hot start cycling profile, which starts with an initial denaturation at 96 °C for 4 min, followed by 25 cycles of amplification at 96 °C/30 s, 64–69.9 °C/30 s, 72 °C/1 kb min−1, and a final extension at 72 °C for 16 min. PCR products were gel purified and sequenced from both ends at Macrogen Corporation, Korea.
Isolation of AaxmCesA1 Promoter Region
The promoter sequence was isolated using the genome walking approach described by Siebert et al. (1995). Two primers, CesAPr2: 5′ ATACGACCCAGCCACCATTCCCACAGT and CesANPr2: 5′ GAAGAAGACAGTGAGAGAGATTTGGAGC were designed using Primer Premier 5 software. Genomic DNA was extracted from the young leaf of an individual Acacia hybrid (AaHy113F1) using the CTAB method (Doyle and Doyle 1990). Isolated genomic DNA was digested with six restriction enzymes: BsrBI, NruI, HpaI, BstZ17I, FspI and SnaBI (New England Biolabs, Hitchin, UK) to produce blunt-end fragments that were then ligated to genome walker adapter. Primary PCR was performed with primer CesAPr2, and then the 10x diluted primary PCR product was used as template for the secondary ‘nested’ PCR with primer CesANPr2. Each PCR reaction contained a final concentration of 1× PCR buffer, 1.5 mM MgCl2, 0.4 μM of each adaptor and gene specific primer and 2 U Hot start Taq, and amplification used a hot-start touch-down PCR cycling profile. Amplification started with a pre-denaturation at 96 °C for 3 min, followed by 7 cycles of amplification at (96 °C/15 s, 72 °C/3 min), then another 20 cycles at (96 °C/15 s, 68 °C/3 min) and ended with a final extension at 68 °C for 15 min. Gel-purified secondary PCR product was sequenced from both ends using ABI PRISM 3730XL DNA analyzer (Applied Biosystems) at Research BioLabs Technologies, Singapore.
Sequence and Structure Analyses
The full-length AaxmCesA1 cDNA sequence isolated was subjected to similarity search using Blast (http://www.ncbi.nlm.nih.gov). A total of 24 CesA protein sequences of the closest homologues were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov) for phylogenetics analysis. The accession numbers for these sequences are: AAT66940.1, XP 003522623.1, AEK31219.1, XP 002515536.1, XP 002324291.1, AAY60847.1, AAY78952.3, ACT16001.1, AAY43218.1, NP 194967.1, AAO25536.1, NP 001054788.1, NP 001105574.1, AAP97497.1, AAF89963.1, XP 002880684.1, NP 180124.1, NP 001051648.1, NP 001105621.1, ADZ16121.1, ADZ16119.1, XP003540527.1, AAT66941.1, and AAY43219.2. In addition to these 24 sequences, selected CesA protein sequences from Arabidopsis (NP 195645.1, NP201279.1, NP 196136.1, Q84JA6, Q8L778, NP 197244.1 and NP 567564 .1), Hordeum vulgare (AY483150, AY483152, AY4831555), Oryza sativa (BAD30574), Zea mays (NP_001104954.1, NP_001104959.1), Gossypium hirsutum (U58283), Bambusa oldhamii (AAY43221.1) and Eucalyptus grandis (AAY60843.1, AAY60845.1, AAY60846.1 and AAY60848.1) were included. Multiple sequences alignment was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/es/cgi-bin/clustalw2) and phylogenetic analysis was carried out using MEGA Version 5 (Tamura et al. 2011). The unrooted phylogenetic tree was constructed using the neighbor-joining method based on distance matrices with 1,000 bootstrap replicates and the Jones-Taylor-Thornton model.
Conserved domains were detected using Conserved Domain software (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), ClustalW2 and Interproscan software (http://www.ebi.ac.uk/Tools/cgi /iprscan.cgi). Promoter sequence was analyzed using Prediction of Plant Promoter (http://www.softberry.com/berry.phtml) and Neural Network Prediction (http://www.fruitly.org/seq_tools/promoter.html) software. The cDNA, introns and promoter sequences amplified from RACE-PCR, primer walking and genome walking respectively, were analyzed using CAP contig assembly (Huang and Madan 1999) and ClustalW2 to predict the AaxmCesA1 full-length gene sequence. The software “Finding 5′, internal and 3′ coding exons” (http://linux.softberry.com/cgi-bin/progrmas/gfind/fex.pl) was applied to predict exon–intron boundaries. The full-length gene sequence was also compared with the full-length gene sequences of other species using ClustalW2 to predict intron and exon positions.
Gene Isoform Analysis
Genomic DNA was extracted from young leaves of an Acacia hybrid individual plant (AaHy113F1) using the CTAB method (Doyle and Doyle 1990). Genomic DNA (15 μg) was digested with two restriction enzymes (BsrBI and BstZ17I; New England Biolabs) at 37 °C for 18 h. The phenol-chloroform purified digested DNA was electrophoresed on a 0.8 % agarose gel in 1x TAE buffer at 25 V for 16 h and transferred to Hybond N + filter membrane (Amersham, Little Chalfont, UK) by downward capillary transfer (Chomczynski 1992) in alkaline buffer (Sambrook and Russell 2001). The blot was hybridized with a 1.6 kb-AaxmCesA probe labeled with 32P radioisotope deoxycytidine 5′-Triphosphate (dCTP) disodium salt using Megaprime DNA Labelling Systems (Amersham), and unincorporated probe was filtered on a Sephadex G-75 column. Hybridization and washing were carried out according to standard procedures described in Sambrook and Russell (2001).
Results
Relative Gene Expression Analysis
The expression levels of AaxmCesA1 in young root, inner bark, young leaf, greenish seedpod and flower tissues were investigated using qPCR. Melt curve analysis showed a single peak for the amplification of AaxmCesA1 gene denoting PCR amplification specificity. Direct sequencing of the qPCR products also confirmed the originality of the amplified products. The highest level of transcript abundance was observed in young root tissue whereas the lowest was evident in inner bark tissue. The expression levels of the AaxmCesA1 gene in the Acacia hybrid were found in a decreasing pattern from leaf, seed pod to flower (Fig. 1). However, statistical analysis using independent t-test at 95 % confidence interval shows that the differences in the expression levels between inner bark tissue and all other tissues studied were not statistically significant with the exception of young root tissue.
Full-length AaxmCesA1 cDNA
The isolated full-length cDNA of the AaxmCesA1 gene is 3,835 bp long, which includes the 3′ UTR (437 bp) and 5′UTR (212 bp) regions. The predicted open reading frame is 1,062 residues in length and encodes a protein of 146 kDa with a predicted pI of 8.84. Multiple alignment analysis performed on the deduced amino acid sequences of the Acacia hybrid and CesA protein sequences of other species revealed a high degree of amino acid similarity throughout most of the sequences length, diverging mostly at the N and C-terminal ends and hypervariable regions (HVR). Although the various CesA proteins from different species varied in their degree of sequence similarity, a conserved motif (D, D, D, QXXRW) was found in all sequences analyzed (Fig. 2). The first, second, third D and QXXRW were found at positions 710, 758, 891 and 929 residues of the translated protein sequence of the Acacia hybrid, respectively.
Comparison of the partial amino acid sequence of Acacia hybrid (labelled FLcDNAaxm) with other plant CesAs using ClustalW. The conserved amino acids/motifs D, D, D, and QXXRW (highlighted in yellow) are found in all the CesAs of the different species. Identical amino acid residues are indicated with asterisks. Accession numbers for the sequences are as follows: E. grandis (gb|AAY60847.1), B. nivea (gb|AAY43218.1), B. oldhamii (gb|aay43218.1), P. tremuloides (gb|AAO25536.1), Z. mays (ref|NP_001105574.1), S. tuberosum (gb|AAP97497.1), A. thaliana (NP_19496.1), A. mangium (gb|AAT66940.1)
Two HVRs were also predicted from the deduced amino acid sequence of AaxmCesA1. HVRI was found before the first aspartic acid in Domain A and HVRII between the second aspartic acid in Domain A and third aspartic acid in Domain B (Fig. 3). HVRI was predicted at positions 201–330, and HVRII at positions 760–830. Other domains, including pfam cellulose_synt, which was identified in the region between residues 394 and 1126, glycosyltransferase family_2 at residues 882–1020, COG1215 (glycosyltransferase) at residues 882–1048 and bcsA at residues 880–1011, were found in the deduced amino acid sequence of the Acacia hybrid. Interpro Scan performed on the full-length AaxmCesA1 cDNA identified additional domains, namely zinc fingers (RING/FYVE/PHD-type) and transmembrane regions. Two putative zinc finger domains were predicted at residues 82–160 and 161–200 in the Acacia hybrid amino acid sequence. Two transmembrane domains were found in the amino terminal region and another six in the carboxy terminal region. The first two transmembrane domains were located before the beginning of the globular domain A at positions 361–381 and 390–410. The other six transmembrane domains were located just after globular domain B at positions 933–953, 963–983, 1002–1020, 1035–1053, 1116–1138 and 1249–1269.
Simplified schematic representation of the major regions of the AaxmCesA1 protein. Horizontal shading Hypervariable regions (HVRI, HRVII), vertical shading putative transmembrane regions, D 1 –D 3 conserved aspartic residues, diagonal shading conserved QXXRW motif, black vertical bar zinc finger domain
Blast X analysis revealed significant matches between the deduced AaxmCesA1 protein sequence and the CesA protein sequences of A. mangium (AAT66940.1), Eucalyptus spp (AEK31219.1; AAY60847.1), Bohmeria nivea (AAY78952.1), Populus spp (XP_002308657.1; AAO25536.1), Arabidopsis thaliana (NP_194967.1), Bambusa oldhamii (AAY43218.1; AAY43216.1) and Ricinus communis (XP_002515536.1), with E-value = 0 and amino acid identity of more than 70 %. Based on the predictions of Blast N analysis, the AaxmCesA1 cDNA shares 95 % nucleotide sequence similarity with Acacia mangium CesA1 (gb|AY643519.1) at E value = 0. It also exhibits over 75 % nucleotide sequence similarity with Ricinus communis CesA6 (ref|XM002515490.1), Glycine max CesA1 (XP_003522623.1), Populus tremuloides CesA4 (gb|162181.1), Populus trichocarpa CesA (ref|XM_002308621.1), Eucalyptus camaldulensis CesA5 (gb|HQ864587.1) and Arabidopsis thaliana CesA1 (ref|NM_119393.2). The high level of similarities in the nucleotide and protein sequences of these CesA genes suggest an orthologous relationship among them. Blast analyses also suggested a possible role for the AaxmCes1 gene in primary cell wall formation of the Acacia hybrid as most of the matched CesA proteins with known function from other species are related to primary cell wall formation.
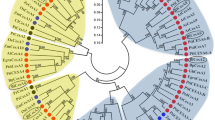
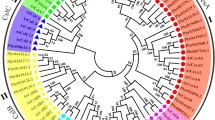
A total of 43 CesA amino acid sequences from other species, in plants ranging from dicots, monocots, angiosperms to gymnosperms, were retrieved from the NCBI website (http://www.ncbi.nlm.nih.gov) and compared with the AaxmCesA1 predicted protein sequence. Figure 4 shows an unrooted tree constructed based on neighbor-joining analysis for the deduced amino acid of AaxmCesA1 with the 43 protein sequences of other plant species. Although not all the CesA genes of these plants have yet been identified, a preliminary conclusion that can be drawn is that, as in most large gene families, orthologous genes are more similar than paralogous genes. Thus, many of the groups in this analysis contain members from plants of both monocot and dicot or angiosperm and gymnosperm lineages.
Unrooted NJ tree with 1,000 bootstrap replicates derived from the alignment of deduced amino acid sequences of AaxmCesA1 with 43 CesA protein sequences (AAT66940.1, XP 003522623.1, AEK31219.1, XP 002515536.1, XP 002324291.1, AAY60847.1, AAY78952.3, ACT16001.1, AAY43218.1, NP 194967.1, AAO25536.1, NP 001054788.1, NP 001105574.1, AAP97497.1, AAF89963.1, XP 002880684.1, NP 180124.1, NP 001051648.1, NP 001105621.1, ADZ16121.1, ADZ16119.1, XP003540527.1, AAT66941.1, AAY43219.2, NP 195645.1, NP201279.1, NP 196136.1, Q84JA6, Q8L778, NP 197244.1, NP 567564 .1, AY483150, AY483152, AY4831555, BAD30574, NP_001104954.1, NP_001104959.1, U58283, AAY43221.1 AAY60843.1, AAY60845.1, AAY60846.1 and AAY60848.1). Aaxm, Acacia hybrid; Am, Acacia mangium; At, Arabidopsis thaliana; Eg, Eucalyptus grandis; Os, Oryza sativa; Zm, Zea mays; Ptr, Populus tremulodes; Gh, Gossypium hirsutum; Bo, Bambusa oldhamii; Pe, Phyllostachys eduli; Rc, Ricinus communis; Hv, Hordeum vulgare L.; Gm, Glycine max; Ec, Eucalyptus camaldulensis; Bn, Bohmeria nivea; St, Solanum tuberocum; Gb, Gossypium barbadense; Gr, Gossypium raimondii; Pt, Populus trichocarpa
Full-Length AaxmCesA1 Gene
A total of nine primer pairs were used to isolate all the intronic regions of the AaxmCesA1 gene. The alignment and assembly of the full-length cDNA sequences obtained from RACE-PCR, and the sequences of introns amplified through primer walking, resulted in a partial gene sequence of 6,545 bp with 13 predicted introns. The gene structure was predicted based on multiple alignment analysis of the full-length AaxmCesA1 gene sequence with the CesA full-length gene sequences of A. thaliana CesA1 (NM119393.2) and CesA10 (128111.2) and full-length cDNA sequences of A. mangium (AY643519) and E. grandis (DQ014509.1). Prediction of these 13 introns was also supported by “Finding 5′, internal and 3′ coding exons” software.
A 1-kb fragment was amplified successfully from Acacia hybrid genomic DNA digested with HpaI using the genome walking approach, which yielded a partial promoter region of 844 bp. Promoter Prediction and Neural Network Prediction software predicted several core promoter elements from the partial nucleotide sequence of the promoter region isolated in this study. The transcription start site (TSS) was found at position 840 bp. Three core elements of the promoter, namely TATA, CAAT and GC boxes were predicted at −32, −53 and −151 bp, respectively, upstream of the TSS of full-length AaxmCesA1 gene. A full-length AaxmCesA1 gene of 7,389 bp was then obtained by assembling the promoter sequence with the partial AaxmCesA1 gene sequence. The isolated AaxmCesA1 consists of 40 % GC and 60 % AT. Blast N analysis of the full-length AaxmCesA1 gene exhibited a significant match to the full-length AtCesA1gene (ref|NC 003075.7) located on chromosome 4 and the AtCesA10 gene located on chromosome 2 of the A. thaliana genome, with E-values of 3e−149 and 0, respectively.
Southern blot analysis using genomic DNA digested with restriction enzymes BstZ17I and Hpal revealed several hybridization bands (Fig. 5). Knowing that most CesA genes in plant species are larger than 4 kb, the hybridization pattern in the Southern analysis suggested that the CesA gene family might consist of up to four isoforms in the Acacia hybrid genome. However, two of these hybridization bands (indicated by arrows in Fig. 5) showed higher intensity than other bands in both digested genomic DNA samples. Based on this, we are confident that there are at least two AaxmCesA isoforms present in the genome of the Acacia hybrid. As the 1.6 kb AaxmCesA1 probe included the HVRII region in its design, sequence variation might cause failure of the probe to hybridize and detect all the different isoforms. This also reflects the possible high sequence variation in the HVRII region of the AaxmCesA genes of the Acacia hybrid but further investigation is necessary.
Discussion
Isolation of a candidate CesA gene is a fundamental step towards understanding the gene functions and mechanisms involved during cellulose biogenesis. Although mutant analyses conducted on Arabidopsis have provided significant information on gene function, we are still far from understanding the complete pathway of cellulose biosynthesis in plants. Furthermore, detailed information on the CesA gene family is available only from a limited number of plants, such as barley (Burton et al. 2004), Arabidopsis (Richmond 2000) and poplar (Djerbi et al. 2004). In the current study, we have isolated and characterized the AaxmCesA1 gene from the Acacia auriculiformis x mangium hybrid for the first time.
Cellulose is the main component in cell wall formation and is found in all plant tissues, but the deposition of cellulose in different cell types varies (Delmer and Amor 1995). In our analysis, qPCR relative gene expression patterns revealed that AaxmCesA1 is expressed in all the tissues investigated but at different levels. The higher levels of transcript abundance observed in young root tissue most probably demonstrates an active involvement of this AaxmCesA1 in the primary cell wall formation of actively dividing young root tissue. Fagard et al. (2000) and Wang et al. (2001) also showed that some CesAs acted early in the process of root hair outgrowth and elongation. The varying expression patterns of this AaxmCesA1 in the five tissues tested here suggested the possibility that more than one CesA is needed for the biosynthesis of cellulose in primary and secondary cell walls in the Acacia hybrid. Previous studies have proposed the involvement of at least three types of CesA isoforms namely, α1, α2 and β, in the spontaneous arrangement of CesAs in the cellulose synthase complex of higher plants (Ding and Himmel 2006; Doblin et al. 2002). In Arabidopsis, a cellulose synthase complex consisting of three unique subunits (AtCesA1, AtCesA3 and AtCesA6) was required for primary cell wall synthesis (Persson et al. 2007). Samuga and Joshi (2002) also showed that two subunits viz., PtrCesA1 and PtrCesA2 are involved in secondary cell wall formation in Populus tremuloides. Another study conducted on the CesA genes of Oryza sativa revealed that OsCesA1, OsCesA3, OsCesA8 and OsCesA4, OsCesA7, OsCesA9 are strongly co-expressed in tissue of typical primary and secondary cell walls respectively. These findings suggest that each set of these OsCesA subunits belongs to the same cellulose synthase complex, which catalyze similar cellulose biosynthesis (Wang et al. 2010; Tanaka et al. 2003). Different types of CesA isoforms have also been found to be co-expressed in barley; HvCesA1, HvCesA2 and HvCesA6 (Burton et al. 2004) and Eucalyptus; EgCesA1, EgCesA2 and EgCesA3 (Ranik and Myburg 2006). Thus, it is very likely that we will find other co-expressing CesA isoforms responsible for the biogenesis of cellulose in either primary or secondary cell wall in the Acacia hybrid.
Based on our Blast analysis, we propose a role in primary cell wall synthesis for the isolated AaxmCesA1, concurring with our relative gene expression study. Blast analyses revealed a high degree of amino acid sequence similarity between the AaxmCesA1 isolated in this study and GmCesA1, EgCesA5 (Ranik and Myburg 2006), OsCesA1 (Wang et al. 2010), BoCesA2 (Chen et al. 2010), AtCesA1 (Desprez et al. 2007) and AmCesA1. The high degree of amino acid sequence similarity suggests that AaxmCesA1 might be orthologous to EgCesA5, AtCesA1, BoCesA2 and OsCesA1, which are all known to be associated with primary cell wall formation. Mutant analysis performed on Arabidopsis in past study showed that a single amino acid substitution in AtCesA1 resulted in a marked reduction of crystalline cellulose synthesis, and the disassembly of cellulose synthase complexes (Arioli et al. 1998). This finding suggests that AtCesA1 is involved in primary cell wall synthesis in Arabidopsis.
The length (3,835 bp) of the AaxmCesA1 cDNA isolated in the current study is similar to that of A. thaliana (3–3.5 kb; Richmond 2000) and poplar (3–3.3 kb; Joshi et al. 2004). The D, D, D, QXXRW signature motif predicted from the deduced amino acid sequence of the Acacia hybrid is a strong predictor for family 2 of β-glycosyltransferases (Campbell et al. 1997). β-Glycosyltransferases can be categorized into processive or non-processive glycosyltransferases depending on the transfer of multiple or single sugar residue to an acceptor molecule. Processive glycosyltransferase is composed of two conserved domains (domains A and B), while non-processive glycosyltransferase contains only domain A (Saxena and Brown 1997). Based on this classification, the AaxmCesA1 isolated in our study is postulated to belong to the processive class because it contains both domains A (consisting two conserved aspartic acid, D) and B (consisting one aspartic acid, D and the QXXRW sequence). The two aspartic acid residues in domain A, and a single aspartic acid residue and the sequence QXXRW in domain B are found to be strictly conserved in all the CesA genes analyzed here. This finding concurs with the analysis performed by Saxena et al. (1994). The distance between the first and second conserved aspartic acid in domain A of the AaxmCesA1 amino acid sequence is 48 residues, while the distance between the second aspartic acid in the domain A with the third aspartic acid in the domain B is 133 residues. A distance of 38 residues is predicted between the third aspartic residues and the QXXRW sequence (Table 2). Saxena et al. (1994) also predicted the distance between the third aspartic acid residue and the QXXRW sequence in domain B to be the most conserved. In contrast, the distance between the two conserved acid aspartic residues in domain A is the most variable, extending from 46 to 120 residues. Saxena and colleagues suggested that these aspartic acid residues might be involved in the catalytic reaction while the QXXRW region was suspected to be responsible for the processivity mechanism.
The prediction of several conserved domains, including the pfam cellulose_synt, glycosyltransferase family_2, COG1215 (glycosyltransferase), bcsA, Interpro zinc finger (RING/FYVE/PHD-type) and transmembrane regions, from the deduced amino acid sequence of the AaxmCesA1 cDNA strongly supports the view that the isolated gene is a cellulose synthase. Cellulose_synt and the glycosyltransferase domains are common to all cellulose synthase genes. Domain bcsA predicted in the AaxmCesA1 protein sequence is also one of the catalytic subunits of cellulose synthase (Wong et al. 1990). Two zinc fingers were also predicted in the N terminus of the AaxmCesA1 deduced amino acid sequence. The prediction of a zinc finger is consistent with the predicted CesA protein features of Arabidopsis (Richmond 2000) and poplar (Joshi et al. 2004). Kurek et al. (2002) identified two putative zinc fingers in the first conserved region of the N terminus of CesA protein in the cotton tree. The location of two zinc fingers within the cytoplasmic N-terminal region of the protein suggested their involvement in protein–protein interactions between cellulose synthase subunits (Kawagoe and Delmer 1997; Kurek et al. 2002; Richmond, 2000). These conserved zinc-finger sequences are suspected to act as redox-regulated multimerization domains that are involved in the assembly of cellulose synthase monomers into rosette complexes.
We also predicted eight transmembrane regions in the AaxmCesA1 amino acid sequence; the distribution of these regions is similar to that of Richmond’s prediction in A. thaliana. According to Richmond (2000), all members of the cellulose synthase superfamily appear to be integral membrane proteins, with one or two transmembrane domains in the amino terminal region, and three to six transmembrane domains in the carboxy terminal region of the protein. This nature of all cellulose synthase superfamily members is crucial, as cellulose is a water-insoluble polymer. Thus, cellulose cannot be synthesized inside the cells but can only be deposited on the external surface of each plant cell. These transmembrane helices are believed to be involved in creating a channel whereby the glucan chain is secreted outside the cell to form new cell walls (Kurek et al. 2002). We also found that the intron–exon organization is highly conserved among most of the CesA genes analyzed in this study. We predicted 13 intron regions in the full-length AaxmCesA1 gene, which is comparable to the arrangement found in A. thaliana (Richmond 2000) and poplar (Joshi et al. 2004). The highly conserved organization of the CesA gene structure across species might reflect its importance for gene functionality in plants.
Analysis of the AaxmCesA1 partial promoter sequence of 840 bp predicted several core elements. These core elements consist of TATA, CAAT and GC boxes, which are generally found in the regulatory regions of promoters and are involved in the regulation of gene expression. Little is known about the transcriptional regulation of this gene family generally, and the Acacia hybrid in particular. A study conducted by Creux et al. (2008) demonstrated that CesA promoters from the Eucalyptus tree are functional in Arabidopsis despite the low sequence similarity between them. Others reported that co-expressed genes harbor conserved transcription factor binding sites (Harmer et al. 2000). Study of the promoter region of AaxmCesA1 is important as it provides an avenue to regulate the expression of AaxmCesA1 in the Acacia hybrid. We aim to isolate the complete promoter region of the AaxmCesA1 and investigate the regulatory mechanism of this gene in our future efforts. Isolation and mutant analyses of the promoter could provide experimental proof for the hypothetical mechanisms orchestrating the transcriptional activity of this gene.
Past investigations conducted on many plant species, including poplar, maize and cotton, have demonstrated that CesA belongs to a multigene family. Desprez et al. (2007) have shown that at least three CesA isoforms are needed for the biosynthesis of cell walls. Different isoforms have been found to be co-expressed in many plant species including rice (Tanaka et al. 2003), Arabidopsis (Taylor et al. 2003), poplar (Joshi et al. 2004) and barley (Burton et al. 2004). Mutant analyses of CesA1, CesA3 and CesA6 showed defects in the primary cell walls of Arabidopsis (Arioli et al. 1998; Fagard et al. 2000). Based on our isoform study, we conjectured that there are other AaxmCesA isoforms apart from AaxmCesA1 that are responsible for the biosynthesis of cellulose in the Acacia hybrid. We predict that at least two isoforms of AaxmCesAs are present in the genome of the Acacia hybrid. The probe used in the isoform analysis was a PCR product amplified from the genomic DNA of the Acacia hybrid flanking region, and includes the conserved aspartic residues D1, D2, D3 and the HVRII. The conserved aspartic residues D1, D2, D3 enable the identification of CesA genes whereas the HVRII helps in differentiating the different isoforms. HVRII was designated as a class specific region (CSR) and has been used to isolate cDNAs encoding CesAs. This is because this region could discriminate each member of the CesA family efficiently (Liang and Joshi 2004; Ranik and Myburg 2006). Therefore, we expect to identify other AaxmCesAs and at the same time distinguish the different isoforms of the AaxmCesA gene family in the Acacia hybrid using this probe. In this current study, we failed to identify a third isoform. Nevertheless, this failure might possibly reflect high sequence variation in the HVRII region of the AaxmCesA genes of the Acacia hybrid. In a previous study conducted on aspen, Samuga and Joshi (2002) concluded that orthologous genes show a higher percentage of identity than paralogous genes. In their work, they found that CesA from the same species exhibits more amino acid sequence variation than between CesA of different species. This variation might explain why we did not detect a third isoform using this probe.
Assessment of gene function through mutant analysis and gene expression studies are important. Although remarkable progress has been made in recent decades, especially in the model plant Arabidopsis, towards unraveling the process of cellulose biosynthesis, many unanswered questions still remain. While mutant analysis can reveal whether a gene is critical for a particular process, it cannot provide us with information on the exact pathways or mechanisms that take place during cellulose deposition. We still have no complete picture of how plants synthesize cellulose during primary and secondary cell wall formation. In the case of the Acacia hybrid, only the AaxmCesA gene has been isolated thus far. Thus, efforts to isolate and investigate all the AaxmCesA genes must continue in order to obtain more a comprehensive insight into the biogenesis of cellulose in the Acacia hybrid. Our current report represents the first step towards achieving this goal.
References
Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, Cork A, Glover J, Redmond J, Williamson RE (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279:717–720
Bowen MR (1981) Acacia mangium. A note on seed collection, handling and storage techniques including some experimental data and information on A. auriculiformis and the probable A. mangium x A. auriculiformis hybrid. Occasional Technical and Scientific Notes, Seed Series Number 3. FAO/UNDP-MAL/78/009
Burn TE, Hocart CH, Birch RJ, Cork AC, Williamson RE (2002) Functional analysis of the cellulose synthase genes CesA1, CesA2 and CesA3 in Arabidopsis. Plant Physiol 129:797–807
Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley—quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134:224–236
Campbell JA, Davies GJ, Bulone VV, Henrissat B (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326:929–939
Chen CY, Hsieh MH, Yang CC, Lin CS, Wang AY (2010) Analysis of the cellulose synthase genes associated with primary cell wall synthesis in Bambusa oldhamii. Phytochemistry 71:1270–1279
Chomczynski P (1992) One-hour downward alkaline capillary transfer for bloting of DNA and RNA. Anal Biochem 201:134–139
Creux NM, Ranik M, Berger DK, Myburg AA (2008) Comparative analysis of orthologous cellulose synthase promoters from Arabidopsis, Populus and Eucalyptus: evidence of conserved regulatory elements in angiosperm. New Phytol 179:722–737
Delmer DP, Amor Y (1995) Cellulose biosynthesis. Plant Cell 7:987–1000
Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Hofte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104(39):15572–15577
Ding SY, Himmel ME (2006) The maize primary cell wall microfibril: a new model derived from direct visualization. J Agric Food Chem 54:597–606
Djerbi S, Lindskog M, Arvested L, Sterky F, Teeri TT (2005) The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 221:739–746
Djerbi S, Aspeborg H, Nilsson P, Sundberg B, Mellerowicz E, Blomqvist K, Teeri TT (2004) Identification and expression analysis of genes encoding putative cellulose synthase (CesA) in the hybrid aspen, Popolua tremula (L.) x P. tremuloides (Michx). Cellulose 11:301–312
Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 42:1407–1420
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, Maureen M, Rayon C, Vernhettes S, Hofte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12:2409–2423
Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113
Hayashi T, Yoshida K, Park YW, Konishi T, Baba K (2005) Cellulose metabolism in plants. Intl Rev Cytol 247:1–34
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Joshi CP, Bhandari S, Ranjan P, Kalluri UC, Liang X, Fujino T, Samuga A (2004) Genomics of cellulose biosynthesis in poplars. New Phytol 164:53–61
Kawagoe Y, Delmer DP (1997) Cotton CelA1 has a LIM-like Zn binding domain in the N-terminal cytoplasmic region. Plant Physiol 114:S-85
Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99:11109–11114
Liang X, Joshi C (2004) Molecular cloning of ten distinct hypervariable regions from the cellulose synthase gene superfamily in aspen trees. Tree Physiol 24:543–550
Maslin BR, Miller JT, Seigler DS (2003) Overview of the generic status of Acacia (Leguminosae: Mimosoideae). Aust Syst Bot 16(1):1–18
McDonald MW, Maslin BR, Butcher PA (2001) Utilisation of Acacias. In: Orchard AE, Wilson AJG (eds) Australian Biological Resources Study. Flora of Australia. CSIRO, Canberra, pp 30–40
Nairn CJ, Haselkorn T (2005) Three loblolly pine CesA genes expressed in developimg xylemare orthologous to secondary cel wall CesA genes of angiosperm. New Phytol 166:907–915
Persson S, Peredez A, Caroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104(39):15566–15571
Ranik M, Myburg AA (2006) Six new cellulose synthase genes from Eucalyptus are associated with primary and secondary cell wall biosynthesis. Tree Physiol 26(5):545–556
Richmond TA (2000) Higher plant cellulose synthases. Genome Biol 1(4):3001.1–3001.6
Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124:495–498
Roberts S, Mouille G, Hofte H (2004) The mechanism and regulation of cellulose synthesis in primary wall: lessons from cellulose-deficient Arabidopsis mutants. Cellulose 11:351–364
Sambrook J, Rusell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY
Samuga A, Joshi C (2002) A new cellulose synthase gene (PtrCesA2) from aspen xylem is orthologous to Arabidopsis AtCesA7 (irx3) gene associated with secondary cell wall synthesis. Gene 296:37–44
Saxena IM, Brown RM (2005) Cellulose Biosynthesis: Current views and evolving concepts. Ann Bot 96:9–21
Saxena IM, Brown RM (1997) Identification of cellulose synthase (s) in higher plants: sequence analysis of processive-glycosyltransferases with the common motif ‘D, D, D35Q(R, Q)XRW’. Cellulose 4:33–49
Saxena IM, Kudlicka K, Okuda K, Brown RM (1994) Characterization of gene in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bact 176(18):5735–5752
Siebert PD, Chencik A, Kellog DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23:1087–1088
Somerville CR (2006) Cellulose synthase in higher plants. Annu Rev Cell Dev Biol 22:53–78
Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H (2003) Three distinct rice cellulose synthase catalytic subunit genes required cellulose synthesis in the secondary wall. Plant Physiol 133:73–83
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interaction among three distinct CESA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100:1450–1455
Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose required for secondary cell wall synthesis. Plant Cell 11:769–780
Tham KC (1976) Introduction to plantation species-Acacia mangium Willd. Proceedings of the 6th Malaysian Forestry Conference, Kuching, Sarawak, Malaysia, pp 153–158
Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9:689–701
Wang HT, Bian MD, Yang ZM, Lin CT, Shi WL (2012) Preliminary functional analysis of the isoforms of OsHsfA2a (Oryza sativa L.) generated by alternative splicing. Plant Mol Biol Rep. doi:10.1007/s11105-012-0471-1
Wang LQ, Guo K, Li Y, Tu YY, Hu HZ, Wang BR, Cui XC, Peng LC (2010) Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol 10:282. doi:10.1007/s11105-012-0471-1
Wang X, Cnops G, Vanderhaeghen R, Block SD, Van Montagu M, Lijsebettens MV (2001) AtCSLD3—acellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol 126:575–586
Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW, Bruner R, Ben-Bassat A, Tal R (1990) Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA 87:8130–8134
Yamada N, Khoo KC, Mohd Nor MY (1990) Sulphate pulping characteristics of Acacia Hybrid, Acacia mangium and Acacia auriculiformis from Sabah. J Trop For Sci 4(3):206–214
Yong SYC, Choong CY, Cheong PL, Pang SL, Nor Amalina R, Harikrishna JA, Mat-Isa MN, Hedley P, Milne L, Vaillancourt R, Wickneswari R (2011) Analysis of ESTs generated from inner bark tissue of an Acacia auriculiformis x Acacia mangium hybrid. Tree Genet Genomes 7(1):143–152
Acknowledgments
This study was funded by the Ministry of Science, Technology and Innovation Malaysia under the Intensified Research Project Area Scheme (Priority Research Grant 01-02-02-0010-PR0003/03-01) and eScience (Project No. 02-01-02-SF0403).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yong, S.Y.C., Wickneswari, R. Molecular characterization of a cellulose synthase gene (AaxmCesA1) isolated from an Acacia auriculiformis x Acacia mangium hybrid. Plant Mol Biol Rep 31, 303–313 (2013). https://doi.org/10.1007/s11105-012-0499-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0499-2