Abstract
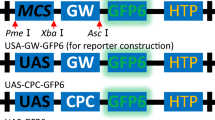
The Agrobacterium tumefaciens VirG response regulator of the VirA/VirG two-component system was adapted to function in tobacco protoplasts. The subcellular localization of VirG and VirA proteins transiently expressed in onion cells was determined using GFP fusions. Preliminary studies using Gal4DBD-VP16 fusions with VirG and Escherichia coli UhpA, and NarL response regulators indicated compatibility of these bacterial proteins with the eukaryotic transcriptional apparatus. A strong transcriptional activator based on tandem activation domains from the Drosophila fushi tarazu and Herpes simplex VP16 was created. Selected configurations of the two-site Gal4-vir box GUS reporters were activated by chimeric effectors dependent on either the yeast Gal4 DNA-binding domain or that of VirG. Transcriptional induction of the GUS reporter was highest for the VirE19-element promoter with both constitutive and wild-type VirG-tandem activation domain effectors. Multiple VirE19 elements increased the reporter activity proportionately, indicating that the VirG DNA binding domain was functional in plants. The VirG constitutive-Q-VP16 effector was more active than the VirG wild-type. In both the constitutive and wild-type forms of VirG, Q-VP16 activated transcription of the GUS reporter best when located at the C-terminus, i.e. juxtaposed to the VirG DNA binding domain. These results demonstrate the possibility of using DNA binding domains from bacterial response regulators and their cognate binding elements in the engineering of plant gene expression.
Graphical Abstract









Similar content being viewed by others
References
Antunes MS, Morey KJ, Tewari-Singh N, Bowen TA, Smith JJ, Webb CT, Hellinga HW, Medford JI (2009) Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol Syst Biol 5:270
Antunes MS, Morey KJ, Smith JJ, Albrecht KD, Bowen TA, Zdunek JK, Troupe JF, Cuneo MJ, Webb CT, Hellinga HW, Medford JI (2011) Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS ONE 6:e16292
Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14:1077–1092
Brencic A, Winans SC (2005) Detection of and response to signals involved in host–microbe interactions by plant-associated bacteria. Microbiol Mol Biol Rev 69:155–1994
Cangelosi GA, Ankenbauer RG, Nester EW (1990) Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci USA 87:6708–6712
Chang CH, Winans SC (1992) Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J Bacteriol 174:7033–7039
Cress WD, Triezenberg SJ (1991) Critical structural elements of the VP16 transcriptional activation domain. Science 251:87–90
Czarnecka-Verner E, Yuan CX, Scharf KD, Englich G, Gurley WB (2000) Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol Biol 43:459–471
Czarnecka-Verner E, Pan S, Salem T, Gurley WB (2004) Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol 56:57–75
Dahl JL, Wei BY, Kadner RJ (1997) Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the UhpT promoter. J Biol Chem 272:1910–1919
Das A, Stachel S, Ebert P, Allenza P, Montoya A, Nester E (1986) Promoters of Agrobacterium tumefaciens Ti-plasmid virulence genes. Nucleic Acids Res 14:1355–1364
Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36:521–528
Fitzpatrick VD, Percival-Smith A, Ingles CJ, Krause HM (1992) Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature 356:610–612
Gao R, Mukhopadhyay A, Fang F, Lynn DG (2006) Constitutive activation of two-component response regulators: characterization of VirG activation in Agrobacterium tumefaciens. J Bacteriol 188:5204–5211
Gao R, Mack TR, Stock AM (2007) Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 32:225–234
Gunsalus RP, Kalman LV, Stewart RR (1989) Nucleotide sequence of the NarL gene that is involved in global regulation of nitrate controlled respiratory genes of Escherichia coli. Nucleic Acids Res 17:1965–1975
Hu X, Zhao J, DeGrado W, Binns A (2013) Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc Natl Acad Sci USA 110:678–683
Hull GA, Devic M (1995) The β-glucuronidase (gus) reporter gene system. Gene fusions; spectrophotometric, fluorometric, and histochemical detection. Method Mol Biol 49:125–141
Jin S, Roitsch T, Ankenbauer RG, Gordon MP, Nester EW (1990a) The VirA protein of Agrobacterium tumefaciens is autophosphorylated and is essential for vir gene regulation. J Bacteriol 172:525–530
Jin SG, Prusti RK, Roitsch T, Ankenbauer RG, Nester EW (1990b) Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J Bacteriol 172:4945–4950
Jin SG, Roitsch T, Christie PJ, Nester EW (1990c) The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J Bacteriol 172:531–537
Jin S, Song Y, Pan SQ, Nester EW (1993) Characterization of a VirG mutation that confers constitutive virulence gene expression in Agrobacterium. Mol Microbiol 7:555–562
Keegan L, Gill G, Ptashne M (1986) Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231:699–704
Kenney LJ (2002) Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol 5:135–141
Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H (2009) Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 284:478–485
Labow MA, Baim SB, Shenk T, Levine AJ (1990) Conversion of the lac repressor into an allosterically regulated transcriptional activator for mammalian cells. Mol Cell Biol 10:3343–3356
Lee K, Dudley MW, Hess KM, Lynn DG, Joerger RD, Binns AN (1992) Mechanism of activation of Agrobacterium virulence genes: identification of phenol-binding proteins. Proc Natl Acad Sci USA 89:8666–8670
Lee YW, Jin S, Sim WS, Nester EW (1995) Genetic evidence for direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 92:12245–12249
Lee YW, Jin S, Sim WS, Nester EW (1996) The sensing of plant signal molecules by Agrobacterium: genetic evidence for direct recognition of phenolic inducers by the VirA protein. Gene 179:83–88
Lin YH, Pierce BD, Fang F, Wise A, Binns AN, Lynn DG (2014) Role of the VirA histidine autokinase of Agrobacterium tumefaciens in the initial steps of pathogenesis. Front Plant Sci 5:195
Looger LL, Dwyer MA, Smith JJ, Hellinga HW (2003) Computational design of receptor and sensor proteins with novel functions. Nature 423:185–190
Mayover TL, Halkides CJ, Stewart RC (1999) Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry 38:2259–2271
Mukhopadhyay A, Gao R, Lynn DG (2004) Integrating input from multiple signals: the VirA/VirG two-component system of Agrobacterium tumefaciens. ChemBioChem 5:1535–1542
Nair GR, Lai X, Wise AA, Rhee BW, Jacobs M, Binns AN (2011) The integrity of the periplasmic domain of the VirA sensor kinase is critical for optimal coordination of the virulence signal response in Agrobacterium tumefaciens. J Bacteriol 193:1436–1448
Narayanan A, Paul LN, Tomar S, Patil DN, Kumar P, Yernool DA (2012) Structure-function studies of DNA binding domain of response regulator KdpE reveals equal affinity interactions at DNA half-sites. PLoS ONE 7:e30102
Pan SQ, Charles T, Jin S, Wu ZL, Nester EW (1993) Preformed dimeric state of the sensor protein VirA is involved in plant—Agrobacterium signal transduction. Proc Natl Acad Sci USA 90:9939–9943
Pazour GJ, Das A (1990a) Characterization of the VirG binding site of Agrobacterium tumefaciens. Nucleic Acids Res 18:6909–6913
Pazour GJ, Das A (1990b) VirG, an Agrobacterium tumefaciens transcriptional activator, initiates translation at a UUG codon and is a sequence-specific DNA-binding protein. J Bacteriol 172:1241–1249
Pazour GJ, Ta CN, Das A (1991) Mutants of Agrobacterium tumefaciens with elevated vir gene expression. Proc Natl Acad Sci USA 88:6941–6945
Pazour GJ, Ta CN, Das A (1992) Constitutive mutations of Agrobacterium tumefaciens transcriptional activator VirG. J Bacteriol 174:4169–4174
Powell BS, Rogowsky PM, Kado CI (1989) VirG of Agrobacterium tumefaciens plasmid pTiC58 encodes a DNA-binding protein. Mol Microbiol 3:411–419
Redrejo-Rodriguez M, Munoz-Espin D, Holguera I, Mencia M, Salas M (2012) Functional eukaryotic nuclear localization signals are widespread in terminal proteins of bacteriophages. Proc Natl Acad Sci USA 109:18482–18487
Roitsch T, Wang H, Jin SG, Nester EW (1990) Mutational analysis of the VirG protein, a transcriptional activator of Agrobacterium tumefaciens virulence genes. J Bacteriol 172:6054–6060
Roitsch T, Jin S, Nester EW (1994) The binding site of the transcriptional activator VirG from Agrobacterium comprises both conserved and specific nonconserved sequences. FEBS Lett 338:127–132
Sadowski I, Ma J, Triezenberg SJ, Ptashne M (1988) Gal4-VP16 is an unusually potent transcriptional activator. Nature 335:563–564
Schrammeijer B, Beijersbergen A, Idler KB, Melchers LS, Thompson DV, Hooykaas PJ (2000) Sequence analysis of the vir-region from Agrobacterium tumefaciens octopine Ti plasmid pTi15955. J Exp Bot 51:1167–1169
Stachel SE, Zambryski PC (1986) VirA and VirG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46:325–333
Tamamoto S, Aoyama T, Takanami M, Oka A (1990) Binding of the regulatory protein VirG to the phased signal sequences upstream from virulence genes on the hairy-root-inducing plasmid. J Mol Biol 215:537–547
Thomas SA, Immormino RM, Bourret RB, Silversmith RE (2013) Nonconserved active site residues modulate CheY autophosphorylation kinetics and phosphodonor preference. Biochemistry 52:2262–2273
Wang R, Brittain MG (2007) The maximum size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett 581:3164–3170
Wang Y, Gao R, Lynn DG (2002) Ratcheting up vir gene expression in Agrobacterium tumefaciens: coiled coils in histidine kinase signal transduction. ChemBioChem 3:311–317
Winans SC, Ebert PR, Stachel SE, Gordon MP, Nester EW (1986) A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci USA 83:8278–8282
Acknowledgments
We thank UF undergraduates Jennifer Cheeseman and Denys Ganyc for excellent technical assistance. We also thank Rob Ferl’s laboratory for help with the confocal microscope. This project was funded by a Grant to L. Ingram and WBG from the United States Defense Advanced Research Project Agency (DARPA) and Office of Naval Research.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
VirA-VirG two-component system of Agrobacterium tumefaciens. VirA is a membrane-imbedded sensor histidine kinase that detects phenolic compounds produced by wounded plant cells (Lee et al. 1995). Detection of acetosyringone ultimately induces a conformational change in VirA resulting in autophosphorylation that triggers a subsequent phosphor residue transfer from His474 of VirA to Asp52 of the VirG response regulator receiver domain (Jin et al. 1990a, 1990b; Lin et al. 2014). The details and identity of other proteins that may be involved in the sensing of phenolics and delivery of that signal to VirA are still unclear (hypothetical proteins X and Y) (Lee et al. 1992). The phosphorylation of VirG facilitates binding of VirG to the vir box promoter elements leading to increased transcription of the vir regulon (Tamamoto et al. 1990). ChvE interacts with the periplasmic domain of VirA and plays an auxiliary role in the induction by increasing the sensitivity of phenolic sensing by VirA (Cangelosi et al. 1990; Nair et al. 2011). The affinity of ChvE for VirA is modulated through ChvE binding to aldose monosaccharaides produced by the host plant (Hu et al. 2013)
Rights and permissions
About this article
Cite this article
Czarnecka-Verner, E., Salem, T. & Gurley, W.B. Adaptation of the Agrobacterium tumefaciens VirG response regulator to activate transcription in plants. Plant Mol Biol 90, 217–231 (2016). https://doi.org/10.1007/s11103-015-0407-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0407-x