Abstract
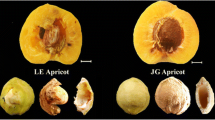
Exocarp color of sand pear is an important trait for the fruit production and has caused our concern for a long time. Our previous study explored the different expression genes between the two genotypes contrasting for exocarp color, which indicated the different suberin, cutin, wax and lignin biosynthesis between the russet- and green-exocarp. In this study, we carried out microscopic observation and Fourier transform infrared spectroscopy analysis to detect the differences of tissue structure and biochemical composition between the russet- and green-exocarp of sand pear. The green exocarp was covered with epidermis and cuticle which was replaced by a cork layer on the surface of russet exocarp, and the chemicals of the russet exocarp were characterized by lignin, cellulose and hemicellulose. We explored differential gene expression between the russet exocarp of ‘Niitaka’ and its green exocarp mutant cv. ‘Suisho’ using Illumina RNA-sequencing. A total of 559 unigenes showed different expression between the two types of exocarp, and 123 of them were common to the previous study. The quantitative real time-PCR analysis supports the RNA-seq-derived gene with different expression between the two types of exocarp and revealed the preferential expression of these genes in exocarp than in mesocarp and fruit core. Gene ontology enrichment analysis revealed divorced expression of lipid metabolic process genes, transport genes, stress responsive genes and other biological process genes in the two types of exocarp. Expression changes in lignin metabolism-related genes were consistent with the different pigmentation of russet and green exocarp. Increased transcripts of putative genes involved the suberin, cutin and wax biosynthesis in ‘Suisho’ exocarp could facilitate deposition of the chemicals and take a role in the mutant trait responsible for the green exocarp. In addition, the divorced expression of ATP-binding cassette transporters involved in the trans-membrane transport of lignin, cutin, and suberin precursors suggests that the transport process could also affect the composition of exocarp and take a role in the regulation of exocarp pigmentation. Results from this study provide a base for the analysis of the molecular mechanism underlying sand pear russet/green exocarp mutation, and presents a comprehensive list of candidate genes that could be used to further investigate the trait mutation at the molecular level.





Similar content being viewed by others
References
Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Lee Y, Geldner N, Fernie AR, Martinoia E (2012) AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol 22(13):1207–1212
Anai T, Koga M, Tanaka H, Kinoshita T, Rahman SM, Takagi Y (2003) Improvement of rice (Oryza sativa L.) seed oil quality through introduction of a soybean microsomal omega-3 fatty acid desaturase gene. Plant Cell Rep 21(10):988–992
Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu XW, Yephremov A, Samuels L (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52(3):485–498
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Chen W, VanOpdorp N, Fitzl D, Tewari J, Friedemann P, Greene T, Thompson S, Kumpatla S, Zheng P (2012) Transposon insertion in a cinnamyl alcohol dehydrogenase gene is responsible for a brown midrib1 mutation in maize. Plant Mol Biol 80(3):289–297
Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F (2009) CYP86B1 is required for very long chain ω-hydroxyacid and α, ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol 150(4):1831–1843
Franke R, McMichael CM, Meyer K, Shirley AM, Cusumano JC, Chapple C (2000) Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J 22(3):223–234
Fu Y, Shen L, Ma K, Wang Y, Ji L, Chen J (1995) Preliminary study of the ‘peel’ structure of cv. ‘Yali’ Pear. In: Proceedings of the thirteenth annual conference of the fruit tree society, Hebei Province. Baoding, Hebei, China
Girard AL, Mounet F, Lemaire-Chamley M, Gaillard C, Elmorjani K, Vivancos J, Runavot JL, Quemener B, Petit J, Germain V, Rothan C, Marion D, Bakan B (2012) Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24(7):3119–3134
Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1)—a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14:545–553
Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R (2008) The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot 59(9):2347–2360
Johansen DA (1940) Plant microtechniques. McGrawHill Book Company Inc, New York
Jové P, Olivella À, Cano L (2011) Study of the variability in chemical composition of bark layers of Quercus suber L. from different production areas. BioResources 6(2):1806–1815
Jung JH, Kim H, Go YS, Lee SB, Hur CG, Kim HU, Suh MC (2011) Identification of functional BrFAD2-1 gene encoding microsomal delta-12 fatty acid desaturase from Brassica rapa and development of Brassica napus containing high oleic acid contents. Plant Cell Rep 30(10):1881–1892
Kaur H, Shaker K, Heinzel N, Ralph J, Gális I, Baldwin IT (2012) Environmental stresses of field growth allow cinnamyl alcohol dehydrogenase-deficient Nicotiana attenuata plants to compensate for their structural deficiencies. Plant Physiol 159(4):1545–1570
Kim D, Hwang J, Shin Y, Shin I, Lee H, Hong S, Kang S (2005) Development of molecular markers linked to several fruit traits in oriental pear. Acta Hortic 671:315–321
Kim J, Jung JH, Lee SB, Go YS, Kim HJ, Cahoon R, Cahoon EB, Markham JE, Suh MC (2013) Arabidopsis 3-ketoacyl-CoA synthase 9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol 162(2):567–580
Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC (2009) Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J 60(3):462–475
Lefever S, Hellemans J, Pattyn F, Przybylski DR, Tayor C, Geurts R, Untergasser A, Vandesompele J (2009) RDML: structured language and reporting guide-lines for real-time quantitative PCR data. Nucleic Acids Res 37:2065–2069
Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106(51):22008–22013
Liu P, Xue C, Wu T-t, Heng W, Jia B, Ye Z, Liu L, Zhu L (2013) Molecular analysis of the processes of surface brown spot (SBS) formation in pear fruit (Pyrus bretschneideri Rehd. cv. Dangshansuli) by de novo transcriptome assembly. PLoS ONE 8(9):e74217. doi:10.1371/journal.pone.0074217
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Malnoy M, Faize M, Venisse J, Geider K, Chevreau E (2005) Expression of viral EPS-depolymerase reduces fire blight susceptibility in transgenic pear. Plant Cell Rep 23:632–638
Mao B, Cheng Z, Lei C, Xu F, Gao S, Ren Y, Wang J, Zhang X, Wang J, Wu F, Guo X, Liu X, Wu C, Wang H, Wan J (2012) Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 235(1):39–52
Marita JM, Vermerris W, Ralph J, Hatfield RD (2003) Variations in the cell wall composition of maize brown midrib mutants. J Agric Food Chem 51(5):1313–1321
Marjamaa K, Kukkola EM, Fagerstedt KV (2009) The role of xylem class III peroxidases in lignification. J Exp Bot 60(2):367–376
Matas AJ, Agustí J, Tadeo FR, Talón M, Rose JK (2010) Tissue-specific transcriptome profiling of the citrus fruit epidermis and subepidermis using laser capture microdissection. J Exp Bot 61(12):3321–3330
Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C (1998) Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA 95(12):6619–6623
Miao YC, Liu CJ (2010) ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci USA 107(52):22728–22733
Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, Aharoni A (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147(2):823–851
Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M (2009) Identification of an Arabidopsis feruloyl-Co A transferase required for suberin synthesis. Plant Physiol 151(3):1317–1328
Panikashvili D, Aharoni A (2011) ABC-type transporters and cuticle assembly: linking function to polarity in epidermis cells. Plant Signal Behav 3(10):806–809
Panikashvili D, Shi JX, Schreiber L, Aharoni A (2009) The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol 151(4):1773–1789
Pereira H (1988) Chemical composition and variability of cork from Quercus suber L. Wood Sci Technol 22:211–218
Pillonel C, Mulder MM, Boon JJ, Forster B, Binder A (1991) Involvement of cinnamyl-alcohol dehydrogenase in the control of lignin formation in Sorghum bicolor L. Moench. Planta 185(4):538–544
Prashant S, Srilakshmi Sunita M, Pramod S, Gupta RK, Anil Kumar S, Rao Karumanchi S, Rawal SK, Kavi Kishor PB (2011) Down-regulation of Leucaena leucocephala cinnamoyl CoA reductase (LlCCR) gene induces significant changes in phenotype, soluble phenolic pools and lignin in transgenic tobacco. Plant Cell Rep 230(12):2215–2231
Provan GJ, Scobbie L, Chesson A (1997) Characterisation of lignin from CAD and OMT deficient Bm mutants of maize. J Sci Food Agric 73(2):133–142
Rowland O, Lee R, Franke R, Schreiber L, Kunst L (2007) The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett 581(18):3538–3544
Ruzin SE (1999) Plant microtechnique and microscopy. Oxford University Press, New York
Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiol 150(2):584–595
Skyba O, Douglas CJ, Mansfield SD (2013) Syringyl-rich lignin renders poplars more resistant to degradation by wood decay fungi. Appl Environ Microbiol 79(8):2560–2571
Storey JD (2002) A direct approach to false discovery rates. J R Stat Soc Ser B (Stat Methodol) 64(3):479–498
Todd J, Post-Beittenmiller D, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17(2):119–130
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Vander Mijnsbrugge K, Beeckman H, De Rycke R, Van Montagu M, Engler G, Boerjan W (2000) Phenylcoumaran benzylic ether reductase, a prominent poplar xylem protein, is strongly associated with phenylpropanoid biosynthesis in lignifying cells. Planta 211(4):502–509
Vermerris W, Sherman DM, McIntyre LM (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61(9):2479–2490
Wagner A, Donaldson L, Kim H, Phillips L, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149(1):370–383
Wagner A, Tobimatsu Y, Goeminne G, Phillips L, Flint H, Steward D, Torr K, Donaldson L, Boerjan W, Ralph J (2013) Suppression of CCR impacts metabolite profile and cell wall composition in Pinus radiata tracheary elements. Plant Mol Biol 81(1–2):105–117
Wang Y, Xu H, Zhang G, Zhu H, Zhang L, Zhang Z, Zhang C, Ma Z (2009) Expression and responses to dehydration and salinity stresses of V-PPase gene members in wheat. J Genet Genomics 36(12):711–720
Wang Y, Dai M, Zhang S, Shi Z (2012) A review on pear bud sport breeding and research progress in mutant mechanisms. J Fruit Sci 29(4):676–682 (in Chinese)
Wang Y, Dai M, Zhang S, Shi Z (2014) Exploring candidate genes for pericarp russet pigmentation of sand pear (Pyrus pyrifolia) via RNA-Seq data in two genotypes contrasting for pericarp color. PLoS ONE 9(1):e83675. doi:10.1371/journal.pone.0083675
Wu P, Tian S, Xu Y (2009) Effects of controlled atmosphere on cell wall and cuticle composition and quality of jujube fruit (cv. Huping). Sci Agric Sin 42(2):619–625
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23(2):396–408
Yan W, Wu G, Liu H, Hou J, Ji A (2009) Observation on exocarp growth of Korean pear during young fruit period. J Shanxi Agric Univ (Nat Sci Ed) 29(1):32–36 (in Chinese)
Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB (2012) A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiol 160(2):638–652
Yu D, Ranathunge K, Huang H, Pei Z, Franke R, Schreiber L, He C (2008) Wax crystal-sparse leaf1 encodes a β-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228(4):675–685
Zhang S, Wu J, Chen H, Gu C, Tao S, Wu J, Zhang S (2011) Identification of differentially expressed genes in a spontaneous mutant of ‘Nanguoli’ pear (Pyrus ussuriensis Maxim) with large fruit. J Hort Sci Biotech 86(6):595–602
Acknowledgments
This study was partially supported by The Special Funds for China “Twelfth Five-Year” National Science and Technology Project for Pear Molecular Breeding and Germplasm Enhancement (2011AA10020602), the Special Funds for China Agriculture Research System (CARS-29-04), The Key Project for New Agricultural Cultivar Breeding in Zhejiang Province, China (2012C112904-2) and The Program for Zhejiang Leading Team of Scientific and Technology Innovation (Grant No. 2009R50033). Fruits of ‘Suisho’ Pear and ‘Niitaka’ Pear used in the study were provided by researcher Yingtao Wang of Fruit Research Institute of Hebei Academy of Agricultural and Forestry Sciences, China. Researcher Yong Li and Dr. Yongbo Wang gave their helps in the materials preparation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yue-zhi Wang and Shujun Zhang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2014_173_MOESM1_ESM.doc
Suppl_Figure 1 Cluster analysis of genes (83 in total) that were differentially expressed between the russet exocarp of ‘Niitaka’ (S3) and the green exocarp of ‘Suisho’ (S4) and verified by the gene expression data for sand pear exocarp russet/green variation. Cluster analysis of genes was performed using Hierarchical cluster analysis. Rows represent differentially expressed genes, columns represent contrast groups. Black and red boxes represent genes showing lower and higher expression level, respectively. The unit of measurement used by Cufflinks to estimate transcript abundance is Fragments Per Kilobase of exon per Million fragments mapped (FPKM). Annotations of genes see Supplemental Table S3 (DOC 77 kb)
11103_2014_173_MOESM2_ESM.doc
Suppl_Figure 2. KEGG pathway of cutin, suberine and wax biosynthesis (ID: ko00073). The arrow indicates the regulation direction of pathways (red) and genes (black) involved in the cutin, suberine and wax biosynthesis in sand pear russet pericarp (expression level of corresponding genes in green exocarp was used as reference) (DOC 303 kb)
11103_2014_173_MOESM3_ESM.doc
Suppl_Figure 3 qRT-PCR analysis of differentially expressed genes detected by the RNA-seq analysis between the exocarps of sand pear cv. ‘Niitaka’ and ‘Suisho’. A1 ~ A3 and B1 ~ B3 represent the exocarp, mesocarp and core of ‘Niitaka’ fruits and ‘Suisho’ fruits, respectively (DOC 1321 kb)
Rights and permissions
About this article
Cite this article
Wang, Yz., Zhang, S., Dai, Ms. et al. Pigmentation in sand pear (Pyrus pyrifolia) fruit: biochemical characterization, gene discovery and expression analysis with exocarp pigmentation mutant. Plant Mol Biol 85, 123–134 (2014). https://doi.org/10.1007/s11103-014-0173-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0173-1