Abstract
Purpose
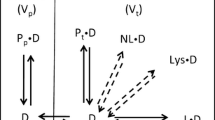
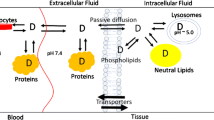
Tissue partitioning is an important component of drug distribution and half-life. Protein binding and lipid partitioning together determine drug distribution.
Methods
Two structure-based models to predict partitioning into microsomal membranes are presented. An orientation-based model was developed using a membrane template and atom-based relative free energy functions to select drug conformations and orientations for neutral and basic drugs.
Results
The resulting model predicts the correct membrane positions for nine compounds tested, and predicts the membrane partitioning for n = 67 drugs with an average fold-error of 2.4. Next, a more facile descriptor-based model was developed for acids, neutrals and bases. This model considers the partitioning of neutral and ionized species at equilibrium, and can predict membrane partitioning with an average fold-error of 2.0 (n = 92 drugs).
Conclusions
Together these models suggest that drug orientation is important for membrane partitioning and that membrane partitioning can be well predicted from physicochemical properties.







Similar content being viewed by others
Abbreviations
- acc:
-
Number of acceptors
- don:
-
Number of donors
- fum :
-
Fraction unbound in microsomal incubation
- fup :
-
Unbound fraction in plasma
- KL :
-
Association constant for drug binding to the lipid
- Kp :
-
Tissue partition constant
- L:
-
Amount of lipid in the tissue
- NO2:
-
Number of NO2 groups
- PBPK:
-
Physiologically based pharmacokinetic models
- pKa,a and pKa,b:
-
pKa values for acids and bases are respectively
- PLS:
-
Partial least squares
- SO:
-
Number of S=O groups
- Vss :
-
Steady-state volume of distribution of a drug
References
Korzekwa K, Nagar S. Compartmental models for apical efflux by P-glycoprotein: part 2-a theoretical study on transporter kinetic parameters. Pharm Res. 2014;31:335–46.
Obach RS. Nonspecific binding to microsomes: impact on scale-up of in vitro intrinsic clearance to hepatic clearance as assessed through examination of warfarin, imipramine, and propranolol. Drug Metab Dispos. 1997;25(12):1359–69.
Tran TH, von Moltke LL, Venkatakrishnan K, Granda BW, Gibbs MA, Obach RS, et al. Microsomal protein concentration modifies the apparent inhibitory potency of CYP3A inhibitors. Drug Metab Dispos. 2002;30(12):1441–5.
Margolis JM, Obach RS. Impact of nonspecific binding to microsomes and phospholipid on the inhibition of cytochrome P4502D6: implications for relating in vitro inhibition data to in vivo drug interactions. Drug Metab Dispos. 2003;31(5):606–11.
Obach RS, Reed-Hagen AE, Krueger SS, Obach BJ, O’Connell TN, Zandi KS, et al. Metabolism and disposition of varenicline, a selective alpha4beta2 acetylcholine receptor partial agonist, in vivo and in vitro. Drug Metab Dispos. 2006;34(1):121–30.
Austin RP, Barton P, Cockroft SL, Wenlock MC, Riley RJ. The influence of nonspecific microsomal binding on apparent intrinsic clearance, and its prediction from physicochemical properties. Drug Metab Dispos. 2002;30(12):1497–503.
Obach RS, Walsky RL, Venkatakrishnan K, Houston JB, Tremaine LM. In vitro cytochrome P450 inhibition data and the prediction of drug-drug interactions: qualitative relationships, quantitative predictions, and the rank-order approach. Clin Pharmacol Ther. 2005;78(6):582–92.
Grime K, Riley RJ. The impact of in vitro binding on in vitro-in vivo extrapolations, projections of metabolic clearance and clinical drug-drug interactions. Curr Drug Metab. 2006;7(3):251.
Obach RS. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: An examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos. 1999;27(11):1350–9.
Riley RJ, McGinnity DF, Austin RP. A unified model for predicting human hepatic, metabolic clearance from in vitro intrinsic clearance data in hepatocytes and microsomes. Drug Metab Dispos. 2005;33(9):1304–11.
Venkatakrishnan K, von Moltke LL, Obach RS, Greenblatt DJ. Microsomal binding of amitriptyline: effect on estimation of enzyme kinetic parameters in vitro. J Pharmacol Exp Ther. 2000;293(2):343–50.
Austin RP, Barton P, Mohmed S, Riley RJ. The binding of drugs to hepatocytes and its relationship to physicochemical properties. Drug Metab Dispos. 2005;33(3):419–25.
Nagar S, Korzekwa K. Commentary: nonspecific protein binding versus membrane partitioning: it is not just semantics. Drug Metab Dispos. 2012;40(9):1649–52.
Hallifax D, Houston JB. Binding of drugs to hepatic microsomes: comment and assessment of current prediction methodology with recommendation for improvement. Drug Metab Dispos. 2006;34(4):724–6. author reply 727.
Poulin P, Haddad S. Microsome composition-based model as a mechanistic tool to predict nonspecific binding of drugs in liver microsomes. J Pharm Sci. 2011;100(10):4501–17.
Poulin P, Haddad S. Hepatocyte composition-based model as a mechanistic tool for predicting the cell suspension: aqueous phase partition coefficient of drugs in in vitro metabolic studies. J Pharm Sci. 2013;102(8):2806–18.
Rodgers T, Leahy D, Rowland M. Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate-to-strong bases. J Pharm Sci. 2005;94(6):1259–76.
Rodgers T, Rowland M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J Pharm Sci. 2006;95(6):1238–57.
Peyret T, Poulin P, Krishnan K. A unified algorithm for predicting partition coefficients for PBPK modeling of drugs and environmental chemicals. Toxicol Appl Pharmacol. 2010;249(3):197–207.
Balaz S. Modeling kinetics of subcellular disposition of chemicals. Chem Rev. 2009;109(5):1793–899.
Naritomi Y, Terashita S, Kimura S, Suzuki A, Kagayama A, Sugiyama Y. Prediction of human hepatic clearance from in vivo animal experiments and in vitro metabolic studies with liver microsomes from animals and humans. Drug Metab Dispos. 2001;29(10):1316–24.
Brown HS, Galetin A, Hallifax D, Houston JB. Prediction of in vivo drug-drug interactions from in vitro data : factors affecting prototypic drug-drug interactions involving CYP2C9, CYP2D6 and CYP3A4. Clin Pharmacokinet. 2006;45(10):1035–50.
Sykes MJ, Sorich MJ, Miners JO. Molecular modeling approaches for the prediction of the nonspecific binding of drugs to hepatic microsomes. J Chem Inf Model. 2006;46(6):2661–73.
Gertz M, Kilford PJ, Houston JB, Galetin A. Drug lipophilicity and microsomal protein concentration as determinants in the prediction of the fraction unbound in microsomal incubations. Drug Metab Dispos. 2008;36(3):535–42.
Kilford PJ, Gertz M, Houston JB, Galetin A. Hepatocellular binding of drugs: correction for unbound fraction in hepatocyte incubations using microsomal binding or drug lipophilicity data. Drug Metab Dispos. 2008;36(7):1194–7.
Heller H, Schaefer M, Schulten K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal phase. J Phys Chem. 1993;97(31):8343–60.
Crivori P, Cruciani G, Carrupt PA, Testa B. Predicting blood–brain barrier permeation from three-dimensional molecular structure. J Med Chem. 2000;43(11):2204–16.
Lukacova V, Natesan S, Peng M, Tandlich R, Wang Z, Lynch S, et al. Structural determinants of drug partitioning in surrogates of phosphatidylcholine bilayer strata. Mol Pharm. 2013;10(10):3684–96.
Li H, Sun J, Sui X, Yan Z, Sun Y, Liu X, et al. Structure-based prediction of the nonspecific binding of drugs to hepatic microsomes. AAPS J. 2009;11(2):364–70.
ACKNOWLEDGMENTS AND DISCLOSURES
This work was partially funded by NIH/NIGMS grants 1R01GM104178 and 1R01GM114369 to KK and SN.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nagar, S., Korzekwa, K. Drug Distribution. Part 1. Models to Predict Membrane Partitioning. Pharm Res 34, 535–543 (2017). https://doi.org/10.1007/s11095-016-2085-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-016-2085-z