Abstract
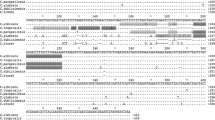
The prevalence of Candida in bloodstream infections (BSIs) has increased. To date, the identification of Candida in BSIs still mainly relies on blood culture and serological tests, but they have various limitations. Therefore, a real-time PCR assay for the detection of Candida from whole blood is presented. The unique primers/probe system was designed on 5.8S rRNA gene (5.8S rDNA) of Candida genus. The analytical sensitivity was determined by numbers of positive PCRs in 12 repetitions. At the concentration of 101 CFU/ml blood, positive PCR rates of 100 % were obtained for C. albicans, C. parapsilosis, C. tropicalis, and C. krusei. The detection rate for C. glabrata was 75 % at 101 CFU/ml blood. The reaction specificity was 100 % when evaluating the assay using DNA samples from clinical isolates and human blood. The maximum CVs of intra-assay and inter-assay for the detection limit were 1.22 and 2.22 %, respectively. To assess the clinical applicability, 328 blood samples from 82 patients were prospectively tested and real-time PCR results were compared with results from blood culture. Diagnostic sensitivity of the PCR was 100 % using as gold standard blood culture, and specificity was 98.4 %. Our data suggest that the developed assay can be used in clinical laboratories as an accurate and rapid screening test for the Candida from whole blood. Although further evaluation is warranted, our assay holds promise for earlier diagnosis of candidemia.

Similar content being viewed by others
References
Luzzaro F, Ortisi G, Larosa M, et al. Prevalence and epidemiology of microbial pathogens causing bloodstream infections: results of the OASIS multicenter study. Diagn Microbiol Infect Dis. 2011;69(4):363–9.
Huttunen R, Attman E, Aittoniemi J, et al. Nosocomial bloodstream infections in a Finnish tertiary care hospital: a retrospective cohort study of 2175 episodes during the years 1999–2001 and 2005–2010. Infect Dis (Lond). 2015;47(1):20–6.
Rodriguez-Creixems M, Munoz P, Martin-Rabadan P, et al. Evolution and aetiological shift of catheter-related bloodstream infection in a whole institution: the microbiology department may act as a watchtower. Clin Microbiol Infect. 2013;19(9):845–51.
Wisplinghoff H, Bischoff T, Tallent SM, et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–17.
Marra AR, Camargo LF, Pignatari AC, et al. Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J Clin Microbiol. 2011;49(5):1866–71.
Wattal C, Raveendran R, Goel N, et al. Ecology of blood stream infection and antibiotic resistance in intensive care unit at a tertiary care hospital in North India. Braz J Infect Dis. 2014;18(3):245–51.
Arendrup MC. Candida and candidaemia. Susceptibility and epidemiology. Dan Med J. 2013;60(11):B4698.
Chen LY, Liao SY, Kuo SC, et al. Changes in the incidence of candidaemia during 2000–2008 in a tertiary medical centre in northern Taiwan. J Hosp Infect. 2011;78(1):50–3.
Nagao M. A multicentre analysis of epidemiology of the nosocomial bloodstream infections in Japanese university hospitals. Clin Microbiol Infect. 2013;19(9):852–8.
Morrell M, Fraser VJ, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother. 2005;49(9):3640–5.
Garey KW, Rege M, Pai MP, et al. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis. 2006;43(1):25–31.
Horvath LL, George BJ, Murray CK, et al. Direct comparison of the BACTEC 9240 and BacT/ALERT 3D automated blood culture systems for Candida growth detection. J Clin Microbiol. 2004;42(1):115–8.
Jamal W, Tamaray G, Pazhoor A, et al. Comparative evaluation of BacT/ALERT 3D and BACTEC systems for the recovery of pathogens causing bloodstream infections. Med Princ Pract. 2006;15(3):223–7.
Gosiewski T, Jurkiewicz-Badacz D, Sroka A, et al. A novel, nested, multiplex, real-time PCR for detection of bacteria and fungi in blood. BMC Microbiol. 2014;14:144.
Wheat LJ. Approach to the diagnosis of invasive aspergillosis and candidiasis. Clin Chest Med. 2009;30(2):367–377, viii.
Persat F, Ranque S, Derouin F, et al. Contribution of the (1 → 3)-beta-d-glucan assay for diagnosis of invasive fungal infections. J Clin Microbiol. 2008;46(3):1009–13.
Lehmann LE, Hunfeld KP, Emrich T, et al. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol. 2008;197(3):313–24.
Taira CL, Okay TS, Delgado AF, et al. A multiplex nested PCR for the detection and identification of Candida species in blood samples of critically ill paediatric patients. BMC Infect Dis. 2014;14:406.
Xafranski H, Melo AS, Machado AM, et al. A quick and low-cost PCR-based assay for Candida spp. identification in positive blood culture bottles. BMC Infect Dis. 2013;13:467.
Foongladda S, Mongkol N, Petlum P, et al. Multi-probe real-time PCR identification of four common Candida species in blood culture broth. Mycopathologia. 2014;177(5–6):251–61.
Lau A, Halliday C, Chen SC, et al. Comparison of whole blood, serum, and plasma for early detection of candidemia by multiplex-tandem PCR. J Clin Microbiol. 2010;48(3):811–6.
Kumar RS, Ramesh S. Novel intein-containing DNA specific primers for rapid identification of Candida glabrata using real-time PCR assays. J Mycol Med. 2014;24(4):337–40.
Loonen AJ, Bos MP, van Meerbergen B, et al. Comparison of pathogen DNA isolation methods from large volumes of whole blood to improve molecular diagnosis of bloodstream infections. PLoS One. 2013;8(8):e72349.
Rodrigues CF, Silva S, Henriques M. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis. 2014;33(5):673–88.
Westh H, Lisby G, Breysse F, et al. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin Microbiol Infect. 2009;15(6):544–51.
Gosiewski T, Brzychczy-Wloch M, Pietrzyk A, et al. Evaluation of the activity of thermostable DNA polymerases in the presence of heme, as a key inhibitor in the real time PCR method in diagnostics of sepsis. Acta Biochim Pol. 2013;60(4):603–6.
Loffler J, Hebart H, Schumacher U, et al. Comparison of different methods for extraction of DNA of fungal pathogens from cultures and blood. J Clin Microbiol. 1997;35(12):3311–2.
Fidel PL Jr, Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev. 1999;12(1):80–96.
Silva S, Negri M, Henriques M, et al. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012;36(2):288–305.
Ruijter JM, Pfaffl MW, Zhao S, et al. Evaluation of qPCR curve analysis methods for reliable biomarker discovery: bias, resolution, precision, and implications. Methods. 2013;59(1):32–46.
Schabereiter-Gurtner C, Selitsch B, Rotter ML, et al. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J Clin Microbiol. 2007;45(3):906–14.
Wellinghausen N, Siegel D, Winter J, et al. Rapid diagnosis of candidaemia by real-time PCR detection of Candida DNA in blood samples. J Med Microbiol. 2009;58(Pt 8):1106–11.
Avni T, Leibovici L, Paul M. PCR diagnosis of invasive candidiasis: systematic review and meta-analysis. J Clin Microbiol. 2011;49(2):6665–70.
Clancy CJ, Nguyen MH. Finding the “missing 50 %” of invasive candidiasis: how nonculture diagnostics will improve understanding of disease spectrum and transform patient care. Clin Infect Dis. 2013;56(9):1284–92.
Acknowledgments
The authors thank Baskova L, et al. (authors of “The Pan-AC assay: a single-reaction real-time PCR test for quantitative detection of a broad range of Aspergillus and Candida species”) for providing the description of procedure of specificity assessment. The study was funded by a college research grant from the Aerospace Center Hospital (YN201322).
Funding
The study was funded by a college research grant from the Aerospace Center Hospital (YN201322).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and Animal Rights
The research involved human participants who donated their blood for preparing simulated candidemia samples and assessing clinical application for the PCR method.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of Aerospace Center Hospital committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Guo, Y., Yang, Jx. & Liang, Gw. A Real-Time PCR Assay Based on 5.8S rRNA Gene (5.8S rDNA) for Rapid Detection of Candida from Whole Blood Samples. Mycopathologia 181, 405–413 (2016). https://doi.org/10.1007/s11046-015-9977-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-015-9977-z