Abstract
Ochroconis bacilliformis, O. phaeophora and O. robusta, three novel species of the melanized genus Ochroconis (Sympoventuriaceae, Venturiales), are described, illustrated and distinguished phenotypically and molecularly from previously described species in the genus Ochroconis. Their potential significance for infection of cold-blooded vertebrates is discussed.
Similar content being viewed by others
Introduction
The genus Ochroconis, typified by O. constricta, was morphologically segregated from a genus with lobed conidia, Scolecobasidium by de Hoog & von Arx [1], for melanized fungi with sympodial conidiogenesis and septate, ellipsoidal conidia which were liberated rhexolytically. Machouart et al. [2] elucidated the general phylogenetic position of the genus by investigating highly conserved genes (nuSSU, nuLSU, mtSSU and RPB2) and found that Ochroconis and its relatives belonged to the order Venturiales, family Sympoventuriaceae. Samerpitak et al. [3] studied species diversity by analyzing more variable genes in addition to the partial ribosomal operon, i.e., the partial coding genes, actin (ACT1), β-tubulin (BT2) and translation elongation factor 1-α (TEF1) and recognized thirteen species in Ochroconis. The authors also introduced the genus Verruconis for a group of thermophilic species such as Ochroconis gallopava, an opportunistic neurotropic pathogen, and its sibling, O. calidifluminalis [4]. The taxonomic status of Scolecobasidium was considered to be doubtful because of ambiguity of the type material, S. terreum [3]. The strict morphological parameters to demarcate the genera were abandoned at the expense of a phylogenetic approach. Species with forked conidia similar to S. terreum were added to Ochroconis on phylogenetic grounds as members of Sympoventuriaceae. Some of the species that were morphologically classified in Scolecobasidium are currently not available for sequencing, and their classification remains unresolved.
Relatively large phylogenetic distances were noted among and within species of Ochroconis and Verruconis [3], which indicated the possible existence of additional, presently unrecognized taxa. Giraldo et al. [5] reported three new species, O. icarus, O. ramosa and O. olivacea, during an investigation of Ochroconis and Verruconis strains originating from clinical samples. The first two are sister species of O. minima, while the latter is close to but significantly different from O. verrucosa. Samerpitak et al. [6] proposed a further new species, O. globalis, which was phylogenetically close to but morphologically different from O. tshawytschae. Crous et al. [7] described a new species, O. macozamiae, as a sister species of O. gamsii which has similar morphology but different genotype.
This article presents a taxonomic study of three Ochroconis strains from various sources. Phenotypic and genotypic characters of the strains were evaluated to define species concepts and delimitations, and three novel Ochroconis species are proposed.
Materials and Methods
Phenotypic Studies
Three Ochroconis strains: CBS 112.97, CBS 206.96 and CBS 100442 (Table 1), were cultured on oatmeal (OA) and malt extract agars (MEA) and incubated at 24 °C for 21 days. Morphological observations were carried out as described by Samerpitak et al. [3, 6]. To investigate the optimal temperature for growth, all strains were grown on MEA incubated for 3 weeks at temperatures varying from 4 to 40 °C with 3 °C intervals. Colony diameters were measured after 3, 7, 11, 14, 18 and 21 days.
Phylogeny
Three Ochroconis strains: CBS 112.97, CBS 206.96 and CBS 100442, including twenty-seven type and reference strains of Ochroconis, Verrconis and neighboring genera (Table 1) were included in phylogenetic analyses. DNA extraction was performed [8–10], three markers, viz. nuSSU, nuLSU and ITS, were amplified by PCR using primers and conditions as shown in Table 2, and sequencing was performed by Big Dye Terminator Cycle Sequencing RR mix protocol (Applied Biosystems). BioNumerics v. 4.61 (Applied Maths, Sint-Martens-Latem, Belgium) was employed for first iterative alignments. Sequences of nuLSU, nuSSU and ITS were aligned with the web-based program Muscle (www.ebi.ac.uk/Tools/msa/muscle). Sequence alignments were adjusted using BioEdit v. 7.0.5.2. Guanine–cytosine content (G+C %) of ITS was calculated using BioEdit v. 7.0.5.2. Sequences were concatenated [3]. Nuclear ribosomal gene analyses were performed in Mega6 [18] using maximum likelihood (ML) with Tamura–Nei and GTR+I as the best model with 1000 bootstrap replicates, and maximum parsimony (MP) with 1000 bootstrap replicates was also carried out. These phylogenetic analyses were supported by the Bayesian approach with MrBayes v. 3.1.2 from the CIPRES Science Gateway [19]. Two parallel runs of 5,000,000 generations were done with a sampling frequency of 1000 trees. A burnin tree sample of 10 % was discarded. The presented tree was obtained with ML approach. Tree reconstruction, visualization and editing were done with TreeView v. 1.6.6, FigTree v. 1.1.2 and Mega6.
Results
The ITS characters of the three investigated strains deviated significantly from those of known species, both in length and in G+C %: CBS 112.97 had 603 bp, 58.04 G+C %; CBS 100442 had 599 bp, 57.43 G+C %; and CBS 206.96 had 554 bp, 61.55 G+C %. This G+C % of CBS 206.96 is the highest comparing to all Ochroconis and Verruconis species (Table 3).
With analyses of partial genes and spacers of the nuclear ribosomal operon, the alignment contained 4103 characters (1656 bp from the SSU, 1040 bp from the ITS, 1407 bp from the LSU) of which 788 were parsimony informative. Applying the algorithms mentioned above, the three investigated strains cluster with Ochroconis spp. in a well-supported clade (95 %ML/99 %MP/1PP) but in different branches (Fig. 1). Single-gene analyses of SSU, ITS or LSU revealed that the three strains invariably remained separate from all known Ochroconis, Verruconis and neighboring species (data not shown). CBS 112.97 and CBS 100442 were strongly supported as sister species (100 %ML/96 %MP/1PP) and found positioned in a cluster containing O. musae, O. constricta, O. minima, O. ramosa and O. icarus (53 %ML/61 %MP/0.99PP). CBS 206.96 clustered as a sister species of O. crassihumicola (84 %ML/96 %MP/1PP) and was paraphyletic to O. cordanae (42 %ML/45 %MP/0.99PP). The significant phylogenetic distances and differential morphological characters underlined that the strains represented three hitherto undescribed species members of the genus Ochroconis.
Mega6 maximum likelihood (ML) tree based on a dataset comprising concatenated gene regions of nuSSU, ITS and nuLSU of Ochroconis, Verruconis and their neighboring genera. The growth profile, associated habitat and pathogenicity information collected from the members of each species have been incorporated into the figure. Numbers at the branches are bootstrap values (%) for Mega6-ML and Mega6-MP, and Bayesian posterior probabilities (PP); ML/MP/PP. Type strains are indicated by (T). A mesophilic, B thermophilic, C oligotrophic, D soil saprophytic, E water associated, F plant associated, G animal associated, H domestic associated, I superficial and cutaneous infections in human, J subcutaneous infection in human, K systemic infection in human, 1 high temperature, 2 including sea water, 3 cold-blooded animal infection, 4 insect colonization, 5 cold-blooded animal infection and insect colonization, 6 warm-blooded animal infection, 7 unpublished data, 8 isolated from clinical specimens, nonsterile site of deep organ; BAL, sputum, etc
Taxonomy
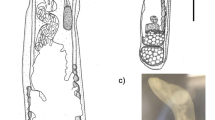
Ochroconis bacilliformis Samerpitak, Gerrits van den Ende, Menken & de Hoog, sp. nov.—MB 810875, Fig. 2.
Etymology: named after its conidial shape.
Specimens examined: Germany, Mülheim, from biofilm on stainless steel in drinking water, 1998, collected by E. Göttlich and identified as Ochroconis constricta. Holotype CBS H-22032 (dried); ex-type culture CBS 100442 (living) = M 37/2.
Description based on CBS 100442 at 24 °C after 3 weeks in darkness.
On OA, colonies 23–24 mm in diameter, moderately expanding, smooth, dry, flat, dark brown; reverse dark brown. On MEA, colonies 17–18 mm in diameter, flat, velvety to floccose, brown; reverse dark brown. Hyphae subhyaline to pale brown, smooth- and thin-walled, 1–2 μm wide; coiled and anastomosing hyphae usually present. Conidiophores mostly arising laterally from vegetative hyphae, conical, flask-shaped to cylindrical, 3–20 × 2–3 µm, pale brown, smooth-walled, with sympodially proliferating conidiogenous cells each bearing one or a few denticles in the apical region; denticles cylindrical, subhyaline, up to 1 μm long. Conidia cylindrical, rounded at both ends or slightly apiculate at the base, 8.8–12.8 × 2.0–2.4 µm, smooth-walled, pale brown, two-celled, becoming verrucose at maturity. Frills remaining visible on denticle and on conidial base. Cardinal temperatures on MEA: minimum at 4 °C, optimum at 18–24 °C, maximum at 30 °C.
Note
The flask-shaped, rather short conical or cylindrical and pale brown conidiophores of Ochroconis bacilliformis are similar to those of O. constricta and O. minima. Its specific character is the conidial shape which is different to Y- to T-shaped or lobate conidia of O. minima, but somewhat similar to that observed in O. constricta, but conidia of O. constricta are shorter but wider (6–12 × 2.5–4.0 µm) and have constriction at the septum [42].
Ochroconis phaeophora Samerpitak, Gerrits van den Ende, Menken & de Hoog, sp. nov.—MB 810876, Fig. 3.
Etymology: named after its dark brown conidiophores.
Specimens examined: Papua New Guinea, Madang, Balek, from leaf in coastal rain forest, 1995, collected by A. Aptroot and A. van Iperen and identified as Ochroconis humicola. Holotype CBS H-22033 (dried); ex-type culture CBS 206.96 (living) = 36599/No. A 165.
Description based on CBS 206.96 at 24 °C after 3 weeks in darkness.
On OA, colonies 40–41 mm in diameter, moderately expanding, smooth, dry, flat, brown to dark brown; reverse dark brown. On MEA, colonies 37–40 mm in diameter, velvety, brown; reverse dark brown with a dark pinkish pigment exuding into the agar. Hyphae subhyaline to pale brown, smooth- to rough- and thick-walled, 1.2–2.8 μm wide; coiled and anastomosing hyphae usually present. Conidiophores mostly arising laterally from vegetative hyphae, erect or flexuous, short- to long-cylindrical with 1–3 (–5) septa, 6–68 × 2.0–2.8 µm, pale brown to dark brown, initially smooth- and thin-walled, rough- and thick-walled at maturity, with sympodially proliferating conidiogenous cells bearing one or more denticles in the apical region; denticles cylindrical, subhyaline, up to 2 μm long. Conidia cylindrical to slightly fusiform, sometimes constricted at the septum, 7.2–11.6 × 2.0–3.6 µm, smooth-walled, pale brown, two-celled, becoming verrucose at maturity. Frills remaining visible on denticle and on conidial base. Cardinal temperatures on MEA: minimum at 4 °C, optimum at 24 °C, maximum at 30 °C.
Note
This species has conidial morphology close to that of O. crassihumicola (CBS 120700), but the conidia of the latter species are longer and wider (7.5–13.0 × 4.2–5.5 µm) and mostly rounded at both ends [34].
Ochroconis robusta Samerpitak, Gerrits van den Ende, Menken & de Hoog, sp. nov.—MB 810874, Fig. 4.
Etymology: named after the well-differentiated conidiophores.
Specimens examined: Spain, from leaf litter of Ouercus ilex, 1996, collected by R.F. Castañeda and identified as Ochroconis humicola. Holotype CBS H-22031 (dried); ex-type culture CBS 112.97 (living) = INIFAT C96/119.
Description based on CBS 112.97 at 24 °C after 3 weeks in darkness.
On OA, colonies 23–24 mm in diameter, dry, flat, dark brown, velvety at the center, smooth at the margin; reverse dark brown. On MEA, colonies 19–21 mm in diameter, velvety with regular shallow radial fissures at the margin, brown; reverse dark brown. Hyphae subhyaline to pale brown, smooth-walled, 1–2 μm wide; coiled and anastomosing hyphae usually present. Conidiophores arising laterally and terminally from vegetative hyphae, erect or flexuous, cylindrical with 1–3 septa, 10–60 × 1.5–2.0 µm, dark brown, smooth- and thick-walled, with sympodially proliferating conidiogenous cells bearing one or more denticles in the apical region; denticles cylindrical, subhyaline, up to 1 μm long. Conidia ellipsoidal to cylindrical, rounded at both ends or slightly apiculated at base, 4.4–6.8 × 2.4–4.0 µm, two-celled, pale brown, smooth-walled, becoming verrucose at maturity. Frills remaining visible on denticle and on conidial base. Cardinal temperatures on MEA: minimum at 4 °C, optimum at 21–24 °C, maximum at 30 °C.
Note
Ochroconis robusta has conidiophores that resemble those of O. humicola; in fact, the strain was originally identified as such. However, the shapes of their conidia are different, and O. humicola produces longer cylindrical conidia (7–15 × 2.5–4.0 µm) [33], while conidia of O. robusta are similar to those observed in O. cordanae, O. globalis and O. anomala. Some subtle morphological characters may help to differentiate such as O cordanae has obovoidal to broadly fusiform conidia [3] and O. anomala produces short chains of conidia [30].
Discussion
Ribosomal gene analyses were sufficient to recognize the three strains, CBS 112.97, CBS 206.96 and CBS 100442, as separate, undescribed species showing strong support of their taxonomic positions within the genus Ochroconis. Phylogenetic analyses of partial coding genes, actin (ACT1), β-tubulin (BT2) and translation elongation factor 1-α (TEF1) were difficult to apply due to high degrees of variability which interfered with alignment over the entire genus. The large phylogenetic distances between species strongly support their identities as novel taxa in the Sympoventuriaceae and suggest that a possible existence of a large number of as yet unrecognized taxa would be waiting to be discovered in unexplored habitats.
With BLAST searches in GenBank, no sequence identical to any of the investigated genes of the new species was encountered. Consequently, although each of the new species is represented by a single strain, their novelty is unambiguous. However, given the morphological variations found upon different culture conditions, identification of these species on the basis of morphology alone remains difficult. Members of the genus have comparable phenotypes despite significant phylogenetic distance between taxa. Nuclear ribosomal sequences of ITS and LSU and even the conserved SSU gene are all usable as diagnostic tools for species identification in Ochroconis. It is remarkable that in the Sympoventuriaceae (including the genera Ochroconis, Verruconis, Veronaeopsis and Sympoventuria), all ribosomal genes, viz. ITS, LSU and the highly conserved SSU, are suitable for identification [3]. This is exceptional in ascomycetous fungi which often share identical ribosomal operons between species, e.g., in Penicillium [43] and Fusarium [44]. ITS sequences with BLAST searches in GenBank directly led to presumptive identification of Ochroconis and Verruconis species. In addition, specific characters in length and G+C % among the pathogenic species (Table 3) provide potentiality to ITS sequences to be developed as tool for definite diagnosis in the medical laboratory.
During the last decade, several reports of new species members in the genus Scolecobasidium—which was considered as of doubtful identification [3]—have appeared such as S. chinense [45], S. qinghaiense [45], S. microsporum [46], S. pallescens [47], S. tuberculatum [48] and S. rostricola [49]. These species all share similar morphology with existing Ochroconis species. Their two-celled conidia may resemble O. robusta and O. phaeophora, but seem to be different in the details and other phenotypic characters. None of them has conidia which are apiculate at both ends, Scolecobasidium chinense has branched conidiophores, S. microsporum has conidia with fine spines at the surface, S. pallescens has caespitose conidiophores, S. qinghaiense has conidia with constricted septa, S. tuberculatum has oblong and tuberculate conidia, and S. rostricola is a fungicolous hyperparasite. The DNA sequences of these Scolecobasidium species are not available in GenBank, and no authentic material was available for study.
Given the limited number of strains per species available, ecological hypotheses are formulated with difficulty. Two species, O. robusta (CBS 112.97) and O. phaeophora (CBS 206.96), are likely to represent plant-associated saprobes, the former having been isolated from leaf litter from Spain and the latter from the leaf under conditions of a tropical rain forest in Papua New Guinea. Ochroconis bacilliformis (CBS 100442) colonized a metal surface submerged in a municipal drinking water network and thus seems to be an oligotrophic saprobe similar to the domestic indoor wet cell colonizers, O. musae, O. constricta, O. globalis [3, 6, 23–25], to the species occurring on cold moist rock, O. anellii [31], O. anomala and O. lascauxensis [30], and also to the waterborne species, O. tshawytschae [3]. Waterborne Ochroconis species have repeatedly been reported as opportunistic agents of disease in cold-blooded animals that complete at least a part of their life cycle in water [22, 28, 29, 50] and may cause superficial infections in humans with impaired blood circulation [5, 29]. Given the identification problems when using phenotypic characteristics, the species introduced in this paper may be likely to have similar abilities.
References
de Hoog GS, von Arx JA. Revision of Scolecobasidium and Pleurophragmium. Kavaka. 1973;1:55–60.
Machouart M, Samerpitak K, de Hoog GS, Gueidan C. A multigene phylogeny reveals that Ochroconis belongs to the family Sympoventuriaceae (Venturiales, Dothideomycetes). Fungal Divers. 2014;65:77–88.
Samerpitak K, Van der Linde E, Choi H-J, Gerrits van den Ende AHG, Machouart M, Gueidan C, de Hoog GS. Taxonomy of Ochroconis, genus including opportunistic pathogens on humans and animals. Fungal Divers. 2014;65:89–126.
Yarita K, Sano A, Samerpitak K, Kamei K, de Hoog GS, Nishimura K. Ochroconis calidifluminalis, a sibling of the neurotropic pathogen O. gallopava, isolated from hot spring. Mycopathologia. 2010;170:21–30.
Giraldo A, Sutton DA, Samerpitak K, de Hoog GS, Wiederhold NP, Guarro J. Gené. Occurrence of Ochroconis and Verruconis species in clinical specimens from the United States. J Clin Microbiol. 2014;52:4189–201.
Samerpitak K, Duarte APM, Attili-Angelis D, Pagnocca FC, Heinrichs G, Rijs AJMM, Alfjorden A, Gerrits van den Ende AHG, Menken SBJ, de Hoog GS. A new species of the oligotrophic genus Ochroconis (Sympoventuriaceae). Mycol Prog. 2015;14:6. doi:10.1007/s11557-015-1023-5.
Crous PW, Shivas RG, Quaedvlieg W, van der Bank M, Zhang Y, Summerell BA, Guarro J, Wingfield MJ, Wood AR, Alfenas AC, Braun U, Cano-Lira JF, García D, Marin-Felix Y, Alvarado P, Andrade JP, Armengol J, Assefa A, den Breeÿen A, Camele I, Cheewangkoon R, De Souza JT, Duong TA, Esteve-Raventós F, Fournier J, Frisullo S, García-Jiménez J, Gardiennet A, Gené J, Hernández-Restrepo M, Hirooka Y, Hospenthal DR, King A, Lechat C, Lombard L, Mang SM, Marbach PAS, Marincowitz S, Marin-Felix Y, Montaño-Mata NJ, Moreno G, Perez CA, Pérez Sierra AM, Robertson JL, Roux J, Rubio E, Schumacher RK, Stchigel AM, Sutton DA, Tan YP, Thompson EH, van der Linde E, Walker AK, Walker DM, Wickes BL, Wong PTW, Groenewald JZ. Fungal Planet description sheets: 214–80. Persoonia. 2014;32:184–306.
Feng P, Lu Q, Gerrits van den Ende AHG, Sun J, Li RY, Xi LY, Vicente VA, Lai W, Lu C, de Hoog GS. Cyphellophora and its relatives in Phialophora: biodiversity and possible role in human infection. Fungal Divers. 2014;65:17–45.
Badali H, Gueidan C, Najafzadeh MJ, Bonifaz A, Gerrits van den Ende AHG, de Hoog GS. Biodiversity of the genus Cladophialophora. Stud Mycol. 2008;61:175–91.
Najafzadeh MJ, Gueidan C, Badali H, Gerrits van den Ende AHG, Xi L, de Hoog GS. Genetic diversity and species delimitation in the opportunistic genus Fonsecaea. Med Mycol. 2009;47:17–25.
White TJ, Bruns T, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for Phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols, a guide to methods and applications. San Diego: Academic; 1990. p. 315–22.
Gargas A, Taylor JW. Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia. 1992;1992(84):589–92.
Hendriks L, Goris A, Neefs J-M, van de Peer Y, Hennebert G, de Wachter R. The nucleotide sequence of the small ribosomal subunit RNA of the yeast Candida albicans and the evolutionary position of the fungi amongst the Eukaryotes. Syst Appl Microbiol. 1989;12:223–9.
de Hoog GS, Göttlich E, Platas G, Genilloud O, Leotta G, van Brummelen J. Evolution, taxonomy and ecology of the genus Thelebolus in Antarctica. Stud Mycol. 2005;51:33–76.
de Hoog GS, Gerrits van den Ende AHG. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–9.
Masclaux F, Guého E, de Hoog GS, Christen R. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J Med Vet Mycol. 1995;33:327–38.
Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–46.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), 14 Nov 2010, New Orleans, LA, 2010, p. 1–8.
Fassatiová O. Notes on the genus Humicola Traaen. II. Ceská Mykol. 1967;21:78–89.
Hao L, Chen C, Zhang R, Zhu M, Sun G, Gleason ML. A new species of Scolecobasidium associated with the sooty blotch and flyspeck complex on banana from China. Mycol Prog. 2013;12:489–95.
Schaumann K, Priebe K. Ochroconis humicola causing muscular black spot disease of Atlantic salmon (Salmo salar). Can J Bot. 1994;72:1629–34.
Lian X, de Hoog GS. Indoor wet cells harbour melanized agents of cutaneous infection. Med Mycol. 2010;48:622–8.
Heinrichs G, Hübner I, Schmidt CK, de Hoog GS, Haase G. Analysis of black fungal biofilms occurring at domestic water taps (I): compositional analysis using tag-encoded FLX amplicon pyrosequencing. Mycopathologia. 2013;175:387–97.
Heinrichs G, Hübner I, Schmidt CK, de Hoog GS, Haase G. Analysis of black fungal biofilms occurring at domestic water taps (II): potential routes of entry. Mycopathologia. 2013;175:399–412.
de Hoog GS. Taxonomy of the Dactylaria complex, IV. Dactylaria, Neta, Subulispora and Scolecobasidium. Stud Mycol. 1985;26:1–60.
Santos DW, Padovan AC, Melo AS, Goncalves SS, Azevedo VR, Ogawa MM, Freitas TV, Colombo AL. Molecular identification of melanised non-sporulating moulds: a useful tool for studying the epidemiology of phaeohyphomycosis. Mycopathologia. 2013;175:445–54.
Doty MS, Slater DW. A new species of Heterobasidium tshawytschae pathogenic on young chinook salmon. Am Midl Nat. 1946;36:663–5.
Ge YP, Lv GX, Shen YN, Li M, Deng SW, de Hoog GS, Samerpitak K, Wei DL. First report of subcutaneous phaeohyphomycosis caused by Ochroconis tshawytschae in an immunocompetent patient. Med Mycol. 2012;50:637–40.
Martin-Sanchez PM, Nováková A, Bastian F, Alabouvette C, Saiz-Jimenez C. Two new species of the genus Ochroconis, O. lascauxensis and O. anomala isolated from black stains in Lascaux Cave, France. Fungal Biol. 2012;116:574–89.
Graniti A. Scolecobasidium anellii n. sp., agenti di annerimenti superficiali di stalattiti. Giorn Bot Ital. 1962;69:360–5.
Matsushima T. Icones Microfungorum a Matsushima lectorum. Kobe: 1975.
Barron GL, Busch LV. Studies on the soil hyphomycete Scolecobasidium. Can J Bot. 1962;40:77–84.
Matsushima T. Microfungi of the Solomon Islands and Papua-New Guinea. Kobe: 1971.
Yarita K, Sano A, Murata Y, Takayama A, Takahashi Y, Takahashi H, Yaguchi T, Ohori A, Kamei K, Miyaji M, Nishimura K. Pathogenicity of Ochroconis gallopava isolated from hot spring in Japan and a review of published reports. Mycopathologia. 2007;164:135–47.
Roy RY, Dwivedi RS, Mishra RR. Two new species of Scolecobasidium from soil. Lloydia. 1962;25:164–6.
Koukol O. Revision of “Septonema ochraceum” revealed three new species of Venturiaceae and Herpotrichiellaceae. Mycol Prog. 2010;9:369–78.
Castañeda RF, Gams W, Saikawa M. Three new conidial fungi (Hyphomycetes) from Cuba. Nova Hedwigia. 1997;64:473–83.
Crous PW, Mohammed C, Glen M, Verkley GJM, Groenewald JZ. Eucalyptus microfungi known from culture. 3. Eucasphaeria and Sympoventuria genera nova, and new species of Furcaspora, Harknessia, Heteroconium and Phacidiella. Fungal Divers. 2007;25:19–36.
Arzanlou M, Groenewald JZ, Gams W, Braun U, Crous PW. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol. 2007;58:57–93.
Sivanesan A, Waller JM. Venturia inaequalis, no. 401. In: CMI descriptions of pathogenic fungi and bacteria. Commonw Mycol Inst, Assoc Biol, Kew, Surrey; 1974. pp. 2.
Abbot EV. Scolecobasidium, a new genus of soil fungi. Mycologia. 1929;19:29–31.
Skouboe P, Taylor JW, Frisvad JC, Lauritsen D, Larsen L, Albæk C, Boysen M, Rossen L. Molecular methods for differentiation of close related Penicillium species. In: Samson RA, Pitt JI, editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam: Harwood Academic Publishers; 2000. p. 179–88.
Wang H, Xiao M, Kong F, Chen S, Dou HT, Sorrell T, Li RY, Xu YC. Accurate and practical identification of 20 Fusarium species by seven-locus sequence analysis and reverse line blot hybridization, and in vitro antifungal susceptibility study. J Clin Microbiol. 2011;49:1890–8.
Pan HQ, Xia HB, Zhang TY. The species of Scolecobasidium from soil in the source areas of Yangtse River and Yellow River. Mycosystema. 2011;31:656–8.
Liu HM, Zhang TY. Two new species of Scolecobasidium. Mycosystema. 2006;25:286–8.
Song W, Yu JF, Zhang TY. Notes on dematiaceous hyphomycetes from soil in Mount Taibai and its surrounding area. Mycosystema. 2008;27:489–96.
Wu YM, Zhang TY. Soil dematiaceous hyphomycetes from Hulunbuir and Xilin Gol Steppe, Inner Mongolia, China. Mycosystema. 2008;27:497–503.
Punithalingam E, Spooner BM. A new fungicolous Scolecobasidium (hyphomycetes) and Caducirostrum gen, nov. (Coelomycetes) from leaf litter in the UK and Italy. Kew Bull. 2011;66:309–24.
de Hoog GS, Guarro J, Gené J, Figueras MJ. Atlas of Clinical Fungi. 4th ed. Utrecht, Reus: CBS-KNAW Fungal Biodiversity Centre, Universitat Rovira i Virgili; 2014.
Acknowledgments
The authors are indebted to Elke Göttlich, André Aptroot, Arien van Iperen and Rafael Castañeda for making material available for study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Taxonomic novelties: Ochroconis bacilliformis Samerpitak, Gerrits van den Ende, Menken & de Hoog, sp. nov., Ochroconis phaeophora Samerpitak, Gerrits van den Ende, Menken & de Hoog, sp. nov., Ochroconis robusta Samerpitak, Gerrits van den Ende, Menken & de Hoog, sp. nov.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Samerpitak, K., Gerrits van den Ende, A.H.G., Menken, S.B.J. et al. Three New Species of the Genus Ochroconis . Mycopathologia 180, 7–17 (2015). https://doi.org/10.1007/s11046-015-9910-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-015-9910-5