Abstract
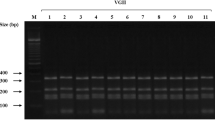
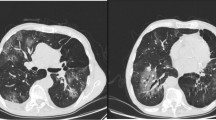
Cryptococcus gattii has becoming more prevalent in temperate climate zones, during the past decades. We describe a C. gattii serotype B infection in an immunocompetent Italian patient with sclerosing cholangitis. The patient traveled once to Eastern Canada and otherwise no other countries than Italy were visited. Molecular analysis revealed that the C. gattii isolate belong to genotype AFLP4/VGI and has mating-type α which is the most common genotype in the Mediterranean environment. The C. gattii strain was found to be closely related, but not identical, to other C. gattii strains from the Mediterranean area.


Similar content being viewed by others
References
Kwon-Chung KJ, Bennett JE. Epidemiological differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984;120:123–30.
Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerg Infect Dis. 2010;16:14–20.
Byrnes EJ III, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199:1081–6.
Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, Hoang L, Leslie MJ, MacDougall L, Magill SS, Morshed MG, Marr KA, Cryptococcus gattii Working Group of the Pacific Northwest. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis. 2009;15:1185–91.
Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, MacDougall L, Boekhout T, Kwon-Chung KJ, Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci USA. 2004;101:17258–63.
Colom MF, Frases S, Ferrer C, Jover A, Andreu M, Reus S, Sánchez M, Torres-Rodríguez JM. First case of human cryptococcosis due to Cryptococcus neoformans var. gattii in Spain. J Clin Microbiol. 2005;43:3548–50.
Colom MF, Hagen F, Gonzalez A, Mellado A, Morera N, Linares C, García DF, Peñataro JS, Boekhout T, Sánchez M. Ceratonia siliqua (carob) trees as natural habitat and source of infection by Cryptococcus gattii in the Mediterranean environment. Med Mycol. 2011. doi:10.3109/13693786.
Montagna MT, Viviani MA, Pulito A, Aralla C, Tortorano AM, Fiore L, Barbuti S. Cryptococcus neoformans var. gattii in Italy. Note II. Environmental investigation related to an autochtonous clinical case in Apulia. J Mycol Med. 1997;7:93–6.
Velegraki A, Kiosses VG, Pitsouni H, Toukas D, Daniilidis VD, Legakis NJ. First report of Cryptococcus neoformans var. gattii serotype B from Greece. Med Mycol. 2001;39:419–22.
Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E, IberoAmerican Cryptococcal Study Group. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–95.
Viviani MA, Cogliati M, Esposto MC, Lemmer K, Tintelnot K, Colom Valiente MF, Swinne D, Velegraki A, Velho R, European Confederation of Medical Mycology (ECMM) Cryptococcosis Working Group. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res. 2006;6:614–9.
Ruma P, Chen SC, Sorrell TC, Brownlee AG. Characterization of Cryptococcus neoformans by random DNA amplification. Lett Appl Microbiol. 1996;23:312–6.
Sorrell TC, Chen SCA, Ruma P, Meyer W, Pfeiffer TJ, Ellis DH, Brownlee AG. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–60.
Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methling K, Viviani MA, Chindamporn A, Sukroongreung S, John MA, Ellis DH, Sorrell TC. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA: a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–9.
Latouche GN, Huynh M, Sorrell TC, Meyer W. PCR restriction fragment length polymorphism analysis of the phospholipase B (PLB1) gene for subtyping of Cryptococcus neoformans isolates. Appl Environ Microbiol. 2003;69:2080–6.
Boekhout T, Theelen B, Diaz M, Fell JW, Hop WCJ, Abeln ECA, Dromer F, Meyer W. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907.
Biswas SK, Wang L, Yokoyama K, Nishimura K. Molecular analysis of Cryptococcus neoformans mitochondrial cytochrome B gene sequences. J Clin Microbiol. 2003;41:5572–6.
Bovers M, Hagen F, Kuramae EE, Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol. 2008;45:400–21.
Diaz MR, Boekhout T, Theelen B, Fell JW. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast Cryptococcus neoformans. Syst Appl Microbiol. 2000;23:535–45.
Hagen F, Boekhout T. The search for the natural habitat of Cryptococcus gattii. Mycopathologia. 2010;170:209–11.
Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, van de Wiele N, Robert V, Currie BJ, Meyer W. Clonality and α-a recombination in the Australian Cryptococcus gattii VGII population: an emerging outbreak in Australia. PLoS One. 2011;6:e16936.
Centers for Disease Control and Prevention (CDC). Emergence of Cryptococcus gattii–Pacific Northwest, 2004–2010. MMWR Morb Mortal Wkly Rep. 2010;59:865-868.
Montagna MT, Tortorano AM, Fiore L, Ingletti AM, Barbuti S. Cryptococcus neoformans var. gattii en Italie. Note I. Premier cas autochtone de méningite de sérotype B chez un sujet VIH positif. J Mycol Med. 1997;7:90–2.
Romeo O, Scordino F, Criseo G. Environmental Isolation of Cryptococcus gattii Serotype B VGI/MATα Strains in Southern Italy. Mycopathologia. 2011;171:423–30.
Hagen F, Illnait-Zaragozi MT, Bartlett KH, Swinne D, Geertsen E, Klaassen CH, Boekhout T, Meis JF. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob Agents Chemother. 2010;54:5139–45.
Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung KJ. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. J Med Mycol. 2009;47:561–70.
Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–4.
Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J Infect Dis. 2005;192:888–92.
Barò T, Torres-Rodriguez JM, De Mendoza MH, Morera Y, Alia C. First identification of autochthonous Cryptococcus neoformans var. gattii isolated from goats with predominantly severe pulmonary disease in Spain. J Clin Microbiol. 1998;36:458–61.
Georgi A, Schneemann M, Tintelnot K, Calligaris-Maibach RC, Meyer S, Weber R, Bosshard PP. Cryptococcus gattii meningoencephalitis in an immunocompetent person 13 months after exposure. Infection. 2009;37:370–3.
Hagen F, van Assen S, Luijckx GJ, Boekhout T, Kampinga GA. Activated dormant Cryptococcus gattii infection in a Dutch tourist who visited Vancouver Island (Canada): a molecular epidemiological approach. Med Mycol. 2010;48:528–31.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Iatta, R., Hagen, F., Fico, C. et al. Cryptococcus gattii Infection in an Immunocompetent Patient from Southern Italy. Mycopathologia 174, 87–92 (2012). https://doi.org/10.1007/s11046-011-9493-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-011-9493-8