Abstract
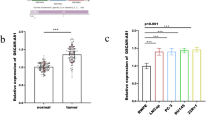
MicroRNAs can regulate many biological functions. miR-122-5p has a tumor suppressor function through different molecular pathways. Also, our second hit, ADAM10, targeted by miR-122-5p, is a major determinant of HER2 shedding causing that trastuzumab cannot bind to HER2 receptors. Therefore, our analysis upon ADAM10 expression and miR-122-5p was a good point to understand molecular mechanism of breast cancer. In our study, we investigated the expression profiles of miR-122-5p and its target ADAM10 in 71 breast cancer patients. Immunohistochemical analysis of ER, PR and HER2 gene products was used to categorize tumors in patients. Expression data and immunohistochemical findings were evaluated to comment on the relationship between miR-122-5p and ADAM10. ADAM10 expression was higher in tumor than that of normal tissue but miR-122-5p expression was lower in tumor than that of normal tissue. The expression pattern in HER2+ patients was reverse of the overall result. It can be explained like that miR-122-5p expression increases especially in HER2+ cancer cell to suppress ADAM10 shedding activity on HER2 receptor. However, increase in expression of tumor suppressor miR-122-5p is not enough to inhibit ADAM10. All in all, we can think miR-122-5p as potential regulator of ADAM10 and trastuzumab resistance. Since if we increase miR-122-5p activity together with trastuzumab administration, then HER2+ breast cancer cells may overcome trastuzumab resistance by inhibiting ADAM10 shedding activity on HER2 receptors and increase the efficiency of trastuzumab.


Similar content being viewed by others
References
Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E (2011) Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer 10:117
World Health Organization (2002) National cancer control programmes policies and managerial guidelines, Geneva. Lancet 365:1727–1741. 2nd Edition
Parkin DM, Fernandez LM (2006) Use of statistics to assess the global burden of breast cancer. Breast J 12(Suppl 1):S70–S80
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen B, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein, Lønning PE, Børresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98(19):10869–10874
Colombo PE, Milanezi F, Weigelt B, Reis-Filho JS (2011) Microarrays in the 2010s: the contribution of microarraybased gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res 13(3):212
Prat A, Perou CM (2011) Deconstructing the molecular portraits of breast cancer. Mol Oncol 5(1):5–23
Horvath A, Pakala SB, Mudvari P, Reddy SD, Ohshiro K, Casimiro S, Pires R, Fuqua SA, Toi M, Costa L, Nair SS, Sukumar S, Kumar R (2013) Novel insights into breast cancer genetic variance through RNA sequencing. Sci Rep 3:2256
Moldovan L, Mitroi A, Petrescu CM, Aschie M (2013) Classification of breast carcinomas according to gene expression profiles. J Med Life 6(1):14–17
Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V (2007) Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 109(9):1721–1728
Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360(8):790–800
Romond EH, Perez EA, Bryant J et al (2005) Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 353(16):1673–1684
Pusztai L, Symmans FW, Hortobagyi GN ( 2005) Development of pharmacogenomic markers to select preoperative chemotherapy for breast cancer. Breast Cancer 12(2):73–85
Zabrecky J, Lam T, McKenzie S, Carney W (1991) The extracellular domain of p185/neu is released from the surface of human breast carcinoma cells, SK-BR-3. J Biol Chem 266:1716–1720
Pupa SM, Menard S, Morelli D, Pozzi B, De Palo G, Colnaghi MI (1993) The extracellular domain of the c-erbB-2 oncoprotein is released from tumor cells by proteolytic cleavage. Oncogene 8:2917–2923
Codony-Servat J, Albanell J, Lopez-Talavera JC, Arribas J, Baselga J (1999) Cleavage of the HER2 ectodomain is a pervanadate-activable process that is inhibited by the tissue inhibitor of metalloproteases-1 in breast cancer cells. Cancer Res 59:1196–1201
Liu PC, Liu X, Li Y, Covington M, Wynn R, Huber R, Hillman M, Yang G, Ellis D, Marando C, Katiyar K, Bradley J, Abremski K, Stow M, Rupar M, Zhuo J, Li YL, Lin Q, Burns D, Xu M, Zhang C, Qian DQ, He C, Sharief V, Weng L, Agrios C, Shi E, Metcalf B, Newton R, Friedman S, Yao W, Scherle P, Hollis G, Burn TC (2006) Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol Ther 5(6):657–664
Yavari R, Adida C, Bray-Ward P, Brines M, Xu T (1998) Human metalloprotease-disintegrin Kuzbanian regulates sympathoadrenal cell fate in development and neoplasia. Hum Mol Genet 7:1161–1167
Fogel M, Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Smirnov A et al (2003) L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 362:869–875
McCulloch DR, Akl P, Samaratunga H, Herington AC, Odorico DM (2004) Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res 10:314–323
Tanida S, Joh T, Itoh K, Kataoka H, Sasaki M, Ohara H et al (2004) The mechanism of cleavage of EGFR ligands induced by inflammatory cytokines in gastric cancer cells. Gastroenterology 127:559–569
Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T et al (2007) Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res 67:7703–7712
Ko SY, Lin SC, Wong YK, Liu CJ, Chang KW, Liu TY (2007) Increase of disintergin metalloprotease 10 (ADAM10) expression in oral squamous cell carcinoma. Cancer Lett 245:33–43
Soundararajan R, Sayat R, Robertson GS, Marignani PA (2009) Triptolide: an inhibitor of a disintegrin and metalloproteinase 10 (ADAM10) in cancer cells. Cancer Biol Ther 8(21):2054–2062
Wolfsberg TG, Primakoff P, Myles DG, White JM (1995) ADAM, a novel family of membrane proteins containing a disintegrin and metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol 131:275–278
Seals DF, Courtneidge SA (2003) The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev 17:7–30
Lunn CA, Fan X, Dalie B, Miller K, Zavodny PJ, Narula SK et al (1997) Purification of ADAM 10 from bovine spleen as a TNFalpha convertase. FEBS Lett 400:333–335
Millichip MI, Dallas DJ, Wu E, Dale S, McKie N (1998) The metallo-disintegrin ADAM10 (MADM) from bovine kidney has type IV collagenase activity in vitro. Biochem Biophys Res Commun 245:594–598
Hattori M, Osterfield M, Flanagan JG (2000) Regulated cleavage of a contact-mediated axon repellent. Science 289:1360–1365
Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, Riedle S et al (2001) Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol 155:661–673
Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W et al (2002) The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet 11:2615–2624
Lemjabbar H, Basbaum C (2002) Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat Med 8:41–46
Yan Y, Shirakabe K, Werb Z (2002) The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol 158:221–226
Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K, Hartmann D, Fahrenholz F, Postina R, Matthews V, Kallen KJ, Rose-John S, Ludwig A (2003) The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 [fractalkine] and regulates CX3CL1-mediated cell-cell adhesion. Blood 102:1186–1195
Nagano O, Murakami D, Hartmann D, de Strooper B, Saftig P, Iwatsubo T et al (2004) Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca(2+) influx and PKC activation. J Cell Biol 165:893–902
Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D et al (2005) ADAM10 cleavage of N-cadherin and regulation of cell–cell adhesion and beta-catenin nuclear signalling. EMBO J 24:742–752
Sanderson MP, Erickson SN, Gough PJ, Garton KJ, Wille PT, Raines EW et al (2005) ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx. J Biol Chem 280:1826–1837
Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S et al (2006) ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol 7:1293–1298
Mochizuki S, Okada Y (2007) ADAMs in cancer cell proliferation and progression. Cancer Sci 98:621–628
Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A et al (2009) MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS ONE 4:e7405
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K, Ren G, Su T, Pan Y, Feng B, Xue Z, Wang X, Fan D (2010) MiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochem Biophys Res Commun 392(3):340–345
Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111–1124
Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T et al (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–689
Esau C, Davis S, Murray SF, Yu XX, Pandey SK et al (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98
Bai S, Nasser MW, Wang B, Hsu SH, Datta J et al (2009) MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem 284:32015–32027
Wang B, Wang H, Yang Z (2012) MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS ONE 7(10):e47053
Wolff AC, Hammond MEH, Hicks DG, Dowsett M et al (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. JCO 31(31):3997–4013
Wu X, Somlo G, Yu Y, Palomares MR, Li AX, Zhou W, Chow A, Yen Y, Rossi JJ, Gao H, Wang J, Yuan YC, Frankel P, Li S, Ashing-Giwa KT, Sun G, Wang Y, Smith R, Robinson K, Ren X, Wang SE (2012) De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med 10:42
Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM et al (2009) A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137:1032–1046
Munagala R, Aqil F, Vadhanam MV, Gupta RC (2013) MicroRNA ‘signature’ during estrogen-mediated mammary carcinogenesis and its reversal by ellagic acid intervention. Cancer Lett 339(2):175–184
Zhou B-BS, Petyon M, He B et al (2006) Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 10:39–50
Fridman JS, Caulder E, Hansbury M et al (2007) Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin Cancer Res 13:1892–1902
Infante J, Burris HA, Lewis N et al (2007) A multicenter phase Ib study of the safety, pharmacokinetics, biological activity and clinical efficacy of INCB7839, a potent and selective inhibitor of ADAM10 and ADAM17. Breast Cancer Res Treat 106(Supp1):S269
Witters L, Scherle P, Friedman S et al (2008) Synergistic inhibition with a dual epidermal growth factor receptor/HER-2/neu tyrosine kinase inhibitor and a disintegrin and metalloprotease inhibitor. Cancer Res 68:7083–7089
Newton RC, Bradley EC, Levy RS et al (2010) Clinical benefit of INCB7839, a potential and selective ADAM inhibitor, in combination with trastuzumab in patients with metastatic HER2-positive breast cancer. J Clin Oncol 28(Suppl; abst 3025):7s
Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J (2014) Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res 42(1):609–621
Hundhausen C, Misztela D, Berkhout TA, et al (2003) The disintegrinlike metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1mediated cellcell adhesion. Blood 102(4):1186–1195
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ergün, S., Ulasli, M., Igci, Y.Z. et al. The association of the expression of miR-122-5p and its target ADAM10 with human breast cancer. Mol Biol Rep 42, 497–505 (2015). https://doi.org/10.1007/s11033-014-3793-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3793-2