Abstract
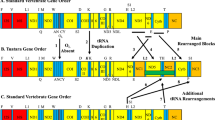
The bryozoan Celleporella has been shown to be composed of multiple, often cryptic, lineages. We sequenced two complete mitochondrial (mt) genomes of the Celleporella hyalina species complex from Wales, UK and Norway (i) to determine genetic divergence at the complete mt genome level, and (ii) to design new molecular markers for examining the interrelationships amongst the major lineages. In addressing (i), we estimated genetic divergence at three levels: (a) nucleotide diversity (π), (b) genome size, and (c) gene order. Genes nad4L, nad6, and atp8 showed the highest levels of divergence, and rrnL, rrnS, and cox1 showed the lowest levels. Inter-genome nucleotide divergence of protein-coding and ribosomal RNA genes, measured as π, was 0.21. The two genomes differed substantially in size, with the Norwegian genome being 2,573 base pairs (bp) longer than the Welsh genome, 17,265 and 14,692 bp, respectively. This difference in size is attributable to long non-coding regions present in the Norwegian genome. Both genomes exhibit similar gene orders, except for the translocation of one transfer RNA (trnA). Considering the high nucleotide diversity, genome size difference and change in gene order, these mt genomes are considered sufficiently divergent to have originated from two distinct species. In addressing (ii) we designed PCR primers that flank the most conserved regions of the genome: 1,300 bp of cox1 and a contiguous 2,000 bp fragment of rrnL + rrnS. The primers have yielded products for tissue from Wales, Norway, New Zealand, Alaska and Chile and should provide useful tools in establishing species- and population-level diversity within the Celleporella complex.






Similar content being viewed by others
References
Mayr E (1942) Systematics and the origin of species, from the viewpoint of a zoologist. Harvard University Press, Cambridge
Willughby RJ, Derham W (1718) Philosophical letters between the late learned Mr. Ray and several of his ingenious correspondents, natives and foreigners: To which are added those of Francis Willughby Esq: the whole consisting of many curious discoveries and improvements in the history of quadrupeds, birds, fishes, insects, plants, fossils, fountains, W and J Innys, London
Winker K (2005) Sibling species were first recognized by William Derham (1718). Auk 122:706–707
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155
Bálint M, Domisch S, Engelhardt CHM, Haase P, Lehrian S, Sauer J, Theissinger K, Pauls SU, Nowak C (2011) Cryptic biodiversity loss linked to global climate change. Nat Clim Change 1:313–318
Lindner A, Govindarajan AF, Migotto AE (2011) Cryptic species, life cycles, and the phylogeny of Clytia (Cnidaria: Hydrozoa: Campanulariidae). Zootaxa 2980:23–36
Baird HP, Miller KJ, Stark JS (2011) Evidence of hidden biodiversity, ongoing speciation and diverse patterns of genetic structure in giant Antarctic amphipods. Mol Ecol 20:3439–3454
Sundberg P, Vodoti ET, Zhou H, Strand M (2009) Polymorphism hides cryptic species in Oerstedia dorsalis (Nemertea, Hoplonemertea). Biol J Linn Soc 98:556–567
Williams S, Apte D, Ozawa T, Kaligis F, Nakano T (2011) Speciation and dispersal along continental coastlines and island arcs in the Indo-West-Pacific turbinid gastropod genus Lunella. Evolution 65:1752–1771
Luttikhuizen PC, Bol A, Cardoso JFMF, Dekker R (2011) Overlapping distributions of cryptic Scoloplos cf. armiger species in the western Wadden Sea. J Sea Res 66:231–237
Hunter RL, Halanych KM (2008) Evaluating connectivity in the brooding brittle star Astrotoma agassizii across the drake passage in the Southern Ocean. J Hered 99:137–148
Xavier JR, Rachello-Dolmen PG, Parra-Velandia F, Schönberg CHL, Breeuwer JAJ, van Soest RWM (2010) Molecular evidence of cryptic speciation in the “cosmopolitan” excavating sponge Cliona celata (Porifera, Clionaidae). Mol Phylogent Evol 56:13–20
Knowlton N (1993) Sibling species in the sea. Annu Rev Ecol Syst 24:189–216
Chimenz Gusso C, Boccia P, Giovannini N (2004) Importance of faunistic and taxonomic studies for a correct analysis of the zoogeography of Mediterranean Bryozoa. Biogeographia 25:93–108
Lee H-J, Kwan Y-S, Kong S-R, Min BS, Seo JE, Won Y-J (2011) DNA barcode examination of Bryozoa (Class: Gymnolaemata) in Korean seawater. Korean J Syst Zool 27:159–163
Dick MH, Herrera-Cubilla A, Jackson JBC (2003) Molecular phylogeny and phylogeography of free-living Bryozoa (Cupuladriidae) from both sides of the Isthmus of Panama. Mol Phylogenet Evol 27:355–371
Schwaninger H (1999) Population structure of the widely dispersing marine bryozoan Membranipora membranacea (Cheilostomata): implications for population history, biogeography, and taxonomy. Mar Biol 135:411–423
Schwaninger H (2008) Global mitochondrial DNA phylogeography and biogeographic history of the antitropically and longitudinally disjunct marine bryozoan Membranipora membranacea (Cheilostomata): another cryptic marine sibling species complex? Mol Phylogenet Evol 49:893–908
Nikulina EA, Hanel R, Schäfer P (2007) Cryptic speciation and paraphyly in the cosmopolitan bryozoan Electra pilosa—impact of the Tethys closing on species evolution. Mol Phylogenet Evol 45:765–776
Nikulina EA (2008) Taxonomy and ribosomal DNA-based phylogeny of the Electra crustulenta species group (Bryozoa: Cheilostomata) with revision of Borg’s varieties and description of Electra moskvikvendi n. sp. from the Western Baltic Sea. Org Divers Evol 8:215–229
Davidson SK, Haygood MG (1999) Identification of sibling species of the bryozoan Bugula neritina that produce different anticancer bryostatins and harbor distinct strains of the bacterial symbiont “Candidatus Endobugula sertula”. Biol Bull 196:273–280
Mackie JA, Keough MJ, Norman JA, Christidis L (2001) Mitochondrial evidence of geographical isolation within Bugula dentata Lamouroux. In: Wyse Jackson PN, Buttler CJ, Spencer Jones ME (eds) Bryozoan Studies 2001, Proceedings of the 12th International Bryozoology Association Conference, AA Balkema Publishers, Dublin, pp 199–206
McGovern TM, Hellberg ME (2003) Cryptic species, cryptic endosymbionts, and geographical variation in chemical defenses in the bryozoan Bugula neritina. Mol Ecol 12:1207–1215
Mackie JA, Keough MJ, Christidis L (2006) Invasion patterns inferred from cytochrome oxidase I sequences in three bryozoans, Bugula neritina, Watersipora subtorquata, and Watersipora arcuata. Mar Biol 149:285–295
Thorpe JP, Beardmore JA, Ryland JS (1978) Genetic evidence for cryptic speciation in the marine bryozoan Alcyonidium gelatinosum. Mar Biol 49:27–32
Thorpe JP, Ryland JS, Beardmore JA (1978) Genetic variation and biochemical systematics in the marine bryozoan Alcyonidium mytili. Mar Biol 49:343–350
Thorpe JP, Ryland JS (1979) Cryptic speciation detected by biochemical genetics in three ecologically important intertidal bryozoans. Estuar Coast Shelf S 8:395–398
Hincks T (1880) A history of the British marine Polyzoa, vol 1. J Van Hoorst, London
Kluge GA (1975) Bryozoa of the Northern Seas of the USSR. Amerind Publishing Co, New Delhi
Morris PA (1980) The bryozoan family Hippothoidae (Cheilostomata–Ascophora) with emphasis on the genus Hippothoa. Mg S AHF 10:1–115
Moyano GHI (1986) Bryozoa marinos Chilenos VI. Cheilostomata Hippothoidae: south eastern Pacific species. Bol Soc Biol Concepción 57:89–135
Hoare K, Goldson AJ, Giannasi N, Hughes RN (2001) Molecular phylogeography of the cosmopolitan bryozoan Celleporella hyalina: cryptic speciation? Mol Phylogenet Evol 18:488–492
Gómez A, Wright PJ, Lunt DH, Cancino JM, Carvalho GR, Hughes RN (2007) Mating trials validate the use of DNA barcoding to reveal cryptic speciation of a marine bryozoan taxon. P Roy Soc Lond B Bio 274:199–207
Gómez A, Hughes RN, Wright PJ, Carvalho GR, Lunt DH (2007) Mitochondrial DNA phylogeography and mating compatibility reveal marked genetic structuring and speciation in the NE Atlantic bryozoan Celleporella hyalina. Mol Ecol 16:2173–2188
Hughes RN, Gómez A, Wright PJ, Moyano HI, Cancino JM, Carvalho GR, Lunt DH (2008) Molecular phylogeny supports division of the ‘cosmopolitan’ taxon Celleporella (Bryozoa; Cheilostomata) into four major clades. Mol Phylogenet Evol 46:369–374
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689
Laslett D, Canbäck B (2008) ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24:172–175
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2011) Geneious v5.4. http://www.geneious.com/
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Katoh K, Kuma K-I, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Yang C-H, Chang H-W, Ho C-H, Chou Y-C, Chuang L-Y (2011) Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS ONE 6:e17729
Oliveira DCSG, Raychoudhury R, Lavrov DV, Werren JH (2008) Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol Biol Evol 25:2167–2180
Jia WZ, Yan HB, Guo AJ, Zhu XQ, Wang YC, Shi WG, Chen HT, Zhan F, Zhang SH, Fu BQ, Littlewood DTJ, Cai XP (2010) Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genomics 11:447
Soroka M (2010) Characteristics of mitochondrial DNA of unionid bivalves (Mollusca: Bivalvia: Unionidae). II. Comparison of complete sequences of maternally inherited mitochondrial genomes of Sinanodonta woodiana and Unio pictorum. Folia Malacol 18:189–209
Podsiadlowski L, Carapelli A, Nardi F, Dallai R, Koch M, Boore JL, Frati F (2006) The mitochondrial genomes of Campodea fragilis and Campodea lubbocki (Hexapoda: Diplura): high genetic divergence in a morphologically uniform taxon. Gene 381:49–61
Inoue JG, Miya M, Lam K, Tay BH, Danks JA, Bell J, Walker TI, Venkatesh B (2010) Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol 27:2576–2586
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Moritz C, Dowling TE, Brown WM (1986) Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu Rev Ecol Syst 18:269–292
Moritz C, Dowling TE, Brown WM (1987) Tandem duplication in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc Natl Acad Sci USA 84:7183–7187
San Mauro D, Gower DJ, Zardoya R, Wilkinson M (2005) A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Biol Evol 23:227–234
Waeschenbach A, Telford MJ, Porter JS, Littlewood DTJ (2006) The complete mitochondrial genome of Flustrellidra hispida and the phylogenetic position of Bryozoa among the Metazoa. Mol Phylogenet Evol 40:195–207
Jang KH, Hwang UW (2009) Complete mitochondrial genome of Bugula neritina (Bryozoa, Gymnolaemata, Cheilostomata): phylogenetic position of Bryozoa and phylogeny of lophophorates within the Lophotrochzoa. BMC Genomics 10:167
Sun M, Wu Z, Shen X, Ren J, Liu X, Liu H, Liu B (2009) The complete mitochondrial genome of Watersipora subtorquata (Bryozoa, Gymnolaemata, Ctenostomata) with phylogenetic consideration of Bryozoa. Gene 439:17–24
Nesnidal MP, Helmkampf M, Bruchhaus I, Hausdorf B (2011) The complete mitochondrial genome of Flustra foliacea (Ectoprocta, Cheilostomata)—compositional bias affects phylogenetic analyses of lophotrochozoan relationships. BMC Genomics 12:572
Sun M, Shen X, Liu H, Liu X, Wu Z, Liu B (2011) Complete mitochondrial genome of Tubulipora flabellaris (Bryozoa: Stenolaemata): the first representative from the class Stenolaemata with unique gene order. Mar Genomics 4:159–165
Boore JL, Collins TM, Stanton D, Daehler LL, Brown WM (1995) Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature 376:163–165
Rokas A, Holland PWH (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15:454–459
Larget B, Kadane JB, Simon DL (2005) A Bayesian approach to the estimation of ancestral genome arrangements. Mol Phylogenet Evol 36:214–223
Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting MF (2009) Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol Biol Evol 26:1607–1617
Sheffield NC, Hiatt KD, Valentine MC, Song HJ, Whiting MF (2010) Mitochondrial genomics in Orthoptera using MOSAS. Mitochondrial DNA 21:87–104
Hyman IT, Ho SYW, Jermiin LS (2007) Molecular phylogeny of Australian helicarionidae euconulidae and related groups (Gastropoda: Pulmonata: Stylommatophora) based on mitochondrial DNA. Mol Phylogenet Evol 45:792–812
Zhou Y, Zhang JY, Zheng RQ, Yu BG, Yang G (2009) Complete nucleotide sequence and gene organization of the mitochondrial genome of Paa spinosa (Anura: Ranoidae). Gene 447:86–96
Waeschenbach A, Taylor PD, Littlewood DTJ (2012) A molecular phylogeny of bryozoans. Mol Phylogenet Evol 62:718–735
Navarrete ZA, Cancino JM, Moyano HI, Hughes RN (2004) Morphological differentiation in the Celleporella hyalina (Linnaeus, 1767) complex (Bryozoa: Cheilostomata) along the Chilean coast. In: Moyano GHI, Cancino JM, Wyse Jackson PN (eds) Bryozoan Studies 2004. Proceedings of the 13th International Bryozoology Association Conference, Concepción, Chile, pp 207–213
Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G (1991) The simple fool’s guide to PCR. University of Hawaii, Department of Zoology and Kewalo Marine Laboratory, Honolulu
Hughes RN (2005) Lessons in modularity: the evolutionary ecology of colonial invertebrates. Sci Mar 69:169–179
Cheetham AH, Jackson JBC, Hayek LAC (1994) Quantitative genetics of bryozoan phenotypic evolution. II. Analysis of selection and random change in fossil species using reconstructed genetic parameters. Evolution 48:360–375
Acknowledgments
We thank the staff at the NHM sequencing facility for their sequencing expertise, Christoffer Schander and Christiane Todt for facilitating the sample collection for the Norwegian genome, and Tim Littlewood and Karin Fehlauer Ale for critical reading of the manuscript. This project was funded by the Natural Environment Research Council, grant number NE/E015298/1, and the Centre for Integrated Research in the Rural Environment, Aberystwyth and Bangor Universities.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2012_1714_MOESM4_ESM.tif
Suppl. Fig. 2 Gel-images of PCR products for a) cox1 (Cellep_cox1F + Cellep_cox1R), product size 1,300 bp, and b) rrnL + rrnS (Cellep_16SR + Cellep_12SF_deg), product size ~ 2000 bp. M = Hyperladder I marker (Bioline); 1 = Wales; 2 = Norway; 3 = New Zealand; 4 = Alaska (Homer); 5 = Alaska (Seldovia); 6 = Chile. (TIFF 3365 kb)
Rights and permissions
About this article
Cite this article
Waeschenbach, A., Porter, J.S. & Hughes, R.N. Molecular variability in the Celleporella hyalina (Bryozoa; Cheilostomata) species complex: evidence for cryptic speciation from complete mitochondrial genomes. Mol Biol Rep 39, 8601–8614 (2012). https://doi.org/10.1007/s11033-012-1714-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1714-9