Abstract
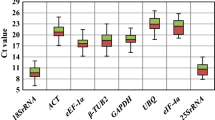
Accurate normalization of gene expression with qRT-PCR depends on the use of appropriate reference genes (RGs) for the species under a given set of experimental conditions. Multiple RGs for gene expression analysis of soybean exposed to heavy metal stress treatment have not been reported in the literature. In this study, we evaluated the expression stability of ten candidate RGs in leaves, roots and stems of two soybean cultivars exposed to cadmium (Cd). Based on the geNorm and NormFinder analysis, ACT3, PP2A, ELF1B and F-box were the most stable RGs in these gene expression studies. In contrast, G6PD, UBC2, TUB, and ELF1A were the most variable ones and should not be used as RGs in these experimental conditions.



Similar content being viewed by others
References
McGrath SP, Zhao FJ, Lombi E (2001) Plant and rhizosphere processes involved in phytoremediation of metal-contaminated soils. Plant Soil 232:207–214
Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212:475–486
Kazantzis G (2004) Cadmium, osteoporosis and calcium metabolism. Biometals 17:493–498
Pandey N, Sharma CP (2002) Effect of heavy metals Co2+, Ni2+ and Cd2+ on growth and metabolism of cabbage. Plant Sci 163:753–758
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plant. Trends Plant Sci 9:490–498
Prasad SM, Zeeshan M (2005) UV-B radiation and cadmium-induced changes in growth, photosynthesis, and antioxidant enzymes of cyanobacterium Plectonema boryanum. Biol Plantarum 49:229–236
Jonak C, Nakagami H, Hirt H (2004) Heavy metal stress. Activation of distinct mitogen-activated protein kinase pathways by copper and cadmium. Plant Physiol 136:3276–3283
Maksymiec W, Krupa Z (2006) The effects of short-term exposition to Cd, excess Cu ions and jasmonate on oxidative stress appearing in Arabidopsis thaliana. Environ Exp Bot 57:187–194
Yeh CM, Chien PS, Huang HJ (2007) Distinct signalling pathways for induction of MAP kinase activities by cadmium and copper in rice roots. J Exp Bot 58:659–671
Jegadeesan S, Yu K, Poysa V, Gawalko E, Morrison MJ, Shi C, Cober E (2010) Mapping and validation of simple sequence repeat markers linked to a major gene controlling seed cadmium accumulation in soybean [Glycine max(L.) Merr]. Theor Appl Genet 121:283–294
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Genome Anal 139:5–17
Bustin SA, Benes V, Nolan T, Pfaffi MW (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34:597–601
Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH (2010) Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol 51:1694–1706
Jian B, Liu B, Bi YR, Hou WS, Wu CX, Han TF (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349
Maroufi A, Bockstaele EV, Loose MD (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Wuytswinkel OV (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Sigh R, Green MR (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259:365–368
Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chuang DM (1996) Evidence that glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. J Neurochem 66:928–935
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Hu RB, Fan CM, Li HY, Zhang QZ, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93
Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1:44–54
Kulcheski FR, Marcelino-Guimaraes FC, Nepomuceno AL, Abdelnoor RV, Margis R (2010) The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal Biochem 406:185–192
Basa B, Solti Á, Sárcári É, Tamás L (2009) Housekeeping gene selection in poplar plants under Cd-stress: comparative study for real-time PCR normalisation. Funct Plant Biol 26:1079–1087
Andersen CL, Jensen JJ, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Ablett GR, Stirling BT, Fischer JD (1999) Westag 97 soybean. Can J Plant Sci 79:371–372
Poysa V, Buzzell RI (2001) AC Hime Soybean. Can J Plant Sci 8:443–444
Arao T, Ae N, Sugiyama M, Takahashi M (2003) Genotypic differences in cadmium uptake and distribution in soybeans. Plant Soil 251:247–253
Redjala T, Sterckeman T, Morel JL (2009) Cadmium uptake by root: contribution of apoplast and high- and low-affinity membrane transport systems. Environ Exp Bot 67:235–242
Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S (2009) Root-to-root Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot 60:2677–2688
Cordoba EM, Die JV, González-Verdejo CI, Nadal S, Román B (2011) Selection of reference genes in Hedysarum coronarium under carious stresses and stages of development. Anal Biochem 409:236–243
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Xu M, Zhang B, Su X, Zhang S, Huang M (2011) Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal Biochem 408:337–339
Huis R, Hawkins S, Neuteling G (2010) Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol 10:87
Smith RD, Walker JC (1996) Plant protein phosphatases. Ann Rev Plant Phys Plant Mol Biol 47:101–125
Filby AL, Tyler CR (2007) Appropriate housekeeping genes for use in expression profiling the effects of environmental estrogens in fish. BMC Mol Biol 8:10
Acknowledgments
The authors thank the Green Farm Ontario and Agriculture and Agri-Food Canada for financial support to K. Yu and V. Poysa and the China Scholarship Council for financial support to Y. Wang.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Yu, K., Poysa, V. et al. Selection of reference genes for normalization of qRT-PCR analysis of differentially expressed genes in soybean exposed to cadmium. Mol Biol Rep 39, 1585–1594 (2012). https://doi.org/10.1007/s11033-011-0897-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0897-9