Abstract
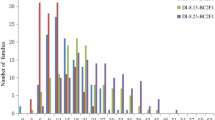
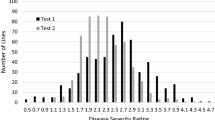
Verticillium wilt (VW) of Upland cotton (Gossypium hirsutum L.) is caused by the soil-borne fungal pathogen Verticillium dahlia Kleb. The availability of VW-resistant cultivars is vital for control of this economically important disease, but there is a paucity of Upland cotton breeding lines and cultivars with a high level of resistance to VW. In general, G. barbadense L. (source of Pima cotton) is more VW-resistant than Upland cotton. However, the transfer of VW resistance from G. barbadense to Upland cotton is challenging because of hybrid breakdown in the F2 and successive generations of interspecific populations. We conducted two replicated greenhouse studies (tests 1 and 2) to assess the heritability of VW resistance to a defoliating V. dahliae isolate and identify genetic markers associated with VW resistance in an Upland cotton recombinant inbred mapping population that has stable introgression from Pima cotton. Disease ratings at the seedling stage on several different days after the first inoculation (DAI) in test 1, as well as the percentages of infected and defoliated leaves at 2 DAI in test 2, were found to be low to moderately heritable, indicating the importance of a replicated progeny test in selection for VW resistance. With a newly constructed linkage map consisting of 882 simple sequence repeat, single nucleotide polymorphism, and resistance gene analog–amplified fragment length polymorphism marker loci, we identified a total of 21 quantitative trait loci (QTLs) on 11 chromosomes and two linkage groups associated with VW resistance at several different DAIs in greenhouse tests 1 and 2. The markers associated with the VW resistance QTLs will facilitate fine mapping and cloning of VW resistance genes and genomics-assisted breeding for VW-resistant cultivars.

Similar content being viewed by others
References
Aguado A, Santos B, Blanco C, Romero F (2008) Study of gene effects for cotton yield and Verticillium wilt tolerance in cotton plant (Gossypium hirsutum L.). Field Crop Res 107:78–86
Anderson MJ, ter Braak CJF (2003) Permutation tests for multi-factorial analysis of variance. J Stat Comput Sim 73:85–113
Barrow JR (1970) Heterozygosity in inheritance of Verticillium wilt tolerance in cotton. Phytopathology 60:301–303
Barrow JR (1973) Genetics of Verticillium tolerance in cotton. In: Ranney CD (ed) Verticillium wilt of cotton. US Agric Res Serv South Reg, S-19:89–97
Bassett D (1974) Resistance of cotton cultivars to verticillium wilt and its relationship to yield. Crop Sci 14:864–867
Bell AA (1992) Verticillium wilt. In: Hillocks RJ (ed) Cotton diseases. CAB International, Wallingford, pp 87–126
Blasingame D, Patel MV (2011) Cotton disease loss estimate committee report. In: Proceedings of Beltwide Cotton Conference, Atlanta, pp 306–308
Blenda A, Fang DD, Rami JM, Garsmeur O, Luo M, Lacape JM (2012) A high density consensus genetic map of tetraploid cotton that integrates multiple component maps through molecular marker redundancy check. PLoS One 7:e45739
Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, Reddy OUK (2005) Mapping of Verticillium wilt resistance genes in cotton. Plant Sci 168:1581–1590
Cantrell RG, Davis DD (2000) Registration of NM24016, an interspecific-derived cotton genetic stock. Crop Sci 40:1208
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
DeVay JE, Weir BL, Wakeman RJ, Stapleton JJ (1997) Effects of Verticillium dahliae infection of cotton plants (Gossypium hirsutum) on potassium levels in leaf petioles. Plant Dis 81:1089–1092
Devey M, Roose M (1987) Genetic analysis of Verticillium wilt tolerance in cotton using pedigree data from three crosses. Theor Appl Genet 74:162–167
Dunnett CW (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121
Fang DD, Yu JZ (2012) Addition of 455 microsatellite marker loci to the high-density Gossypium hirsutum TM-1 x G. barbadense 3-79 genetic map. J Cotton Sci 16:229–248
Fang H, Zhou H, Sanogo S, Flynn R, Percy RG, Hughs SE, Ulloa M, Jones DC, Zhang J (2013) Quantitative trait locus mapping for Verticillium wilt resistance in a backcross inbred line population of cotton (Gossypium hirsutum × Gossypium barbadense) based on RGA-AFLP analysis. Euphytica 194:79–91
Gore MA, Percy RG, Zhang J, Fang DD, Cantrell RG (2012) Registration of the TM-1/NM24016 cotton recombinant inbred mapping population. J Plant Reg 6:124–127
Gore M A, Fang DD, Poland, JA, Zhang J, Percy RG, Fang DD, Cantrell RG, Thyssen G, Lipka AE (2013) Linkage map construction and QTL analysis of agronomic and fiber quality traits in cotton. Plant Genome (in press)
Hayter AJ (1986) The maximum familywise error rate of Fisher’s least significant difference test. J Am Stat Assoc 81:1000–1004
Holland JB, Nyquist WE, Cervantes-Martínez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Jiang F, Zhao J, Zhou L, Guo WZ, Zhang TZ (2009) Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in Upland cotton. Sci China Ser C Life Sci 52:872–884
Kohel R, Richmond T, Lewis C (1970) Texas Marker-1. Description of a genetic standard for Gossypium hirsutum L. Crop Sci 10:670–671
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
McCouch S, Cho Y, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newslett 14:11–13
Mert M, Kurt S, Gencer O, Akiscan Y, Boyaci K, Tok FM (2005) Inheritance of resistance to Verticillium wilt (Verticillium dahliae) in cotton (Gossypium hirsutum L.). Plant Breed 124:102–104
Ning ZY, Zhao R, Chen C, Ai NJ, Zhang X, Zhao J, Mei HX, Wang P, Guo WZ, Zhang TZ (2013) Molecular tagging of a major QTL for broad-spectrum resistance to Verticillium wilt in Upland cotton cultivar Prema. Crop Sci 53:2304–2312
Niu C, Lu Y, Yuan Y, Percy R, Ulloa M, Zhang JF (2011) Mapping resistance gene analogs (RGAs) in cultivated tetraploid cotton using RGA-AFLP analysis. Euphytica 181:65–76
Paplomatas E, Bassett D, Broome J, DeVay J (1992) Incidence of Verticillium wilt and yield losses of cotton cultivars (Gossypium hirsutum) based on soil inoculum density of Verticillium dahliae. Phytopathology 82:1417–1420
Percy RG, Cantrell RG, Zhang JF (2006) Genetic variation for agronomic and fiber properties in an introgressed recombinant inbred population of cotton. Crop Sci 46:1311–1317
Pullman GS, DeVay JE (1982) Epidemiology of Verticillium wilt of cotton: effects of disease development on plant phenology and lint yield. Phytopathology 72:554–559
Roberts CL, Staten G (1972) Heritability of Verticillium wilt tolerance in crosses of American Upland cotton. Crop Sci 12:63–66
SAS Institute (2012) The SAS system for Windows. Release 9.3. SAS Inst, Cary, NC
Smith CW, Cothren JT (1999) Cotton: origin, history, technology, and production. Wiley, New York
USDA-NASS (2013) Crop Production 2012 Summary. January 2013. http://usda01.library.cornell.edu/usda/current/CropProdSu/CropProdSu-01-11-2013.pdf. Accessed 17 March 2013
van Ooijen JW (2006) JoinMap® 4.0, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V, Wageningen
van Ooijen JW (2009) MapQTL® 6, Sofware for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B.V, Wageningen
Verhalen LM, Brinkerhorf LA, Fun KC, Morrison WC (1971) A quantitative genetic study of Verticillium wilt resistance among selected lines of Upland cotton. Crop Sci 11:407–412
Wang HM, Lin ZX, Zhang XL, Chen W, Guo XP, Nie YC, Li YH (2008) Mapping and quantitative trait loci analysis of Verticillium wilt resistance genes in cotton. J Integr Plant Biol 50:174–182
Wheeler TA, Woodward JE (2011) Affect of Verticillium wilt on cultivars in the Southern High Plains of Texas. In: Proceedings of Beltwide Cotton Conference, Atlanta, 4–7 Jan 2011, pp 293–305
Xu S (2003) Theoretical basis of the Beavis effect. Genetics 165:2259–2268
Yang C, Guo WZ, Li GY, Gao F, Lin SS, Zhang TZ (2008) QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci 174:290–298
Zhang JF, Percy RG (2007) Improving Upland cotton by introducing desirable genes from Pima cotton. World Cotton Res Conf (http://wcrc.confex.com/wcrc/2007/techprogram/P1901.HTM)
Zhang JF, Stewart JM (2000) Economical and rapid method for extracting cotton genomic DNA. J Cotton Sci 4:193–201
Zhang JF, Lu Y, Adragna H, Hughs E (2005a) Genetic improvement of New Mexico Acala cotton germplasm and their genetic diversity. Crop Sci 45:2363–2373
Zhang JF, Lu Y, Cantrell RG, Hughs E (2005b) Molecular marker diversity and field performance in commercial cotton cultivars evaluated in the Southwestern USA. Crop Sci 45:1483–1490
Zhang JF, Yuan YL, Niu C, Hinchliffe DJ, Lu YZ, Yu SX, Percy RG, Ulloa M, Cantrell RG (2007) AFLP-RGA markers in comparison with RGA and AFLP in cultivated tetraploid cotton. Crop Sci 47:180–187
Zhang JF, Sanogo S, Flynn R, Baral JB, Bajaj S, Hughs SE, Percy RG (2012) Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 187:147–160
Zhang JF, Fang H, Zhou HP, Sanogo S (2013) Genetics, breeding, and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci (Accepted upon revision)
Acknowledgments
The research was supported by the USDA-ARS, Cotton Incorporated, and New Mexico Agricultural Experiment Station. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Fang, H., Zhou, H., Sanogo, S. et al. Quantitative trait locus analysis of Verticillium wilt resistance in an introgressed recombinant inbred population of Upland cotton. Mol Breeding 33, 709–720 (2014). https://doi.org/10.1007/s11032-013-9987-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9987-9