Abstract
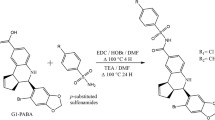
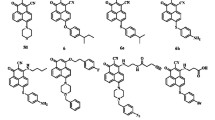
The main aim of this study was to discover small molecule inhibitors against Cathepsin D (CatD) (EC.3.4.23.5), a clinically proven prognostic marker for breast cancer, and to explore the mechanisms by which CatD could be a useful therapeutic target for triple-positive and triple-negative breast cancers (TPBC & TNBC). The crystal structure of CatD at 2.5 Å resolution (PDB: 1LYB), which was complexed with Pepstatin A, was selected for computer-aided molecular modeling. The methods used in our study were pharmacophore modeling and molecular docking. Virtual screening was performed to identify small molecules from an in-house database and a large commercial chemical library. Cytotoxicity studies were performed on human normal cell line HEK293T and growth inhibition studies on breast adenocarcinoma cell lines, namely MCF-7, MDA-MB-231, SK-BR-3, and MDA-MB-468. Furthermore, RT-PCR analysis, in vitro enzyme assay, and cell cycle analysis ascertained the validity of the selected molecules. A set of 28 molecules was subjected to an in vitro fluorescence-based inhibitory activity assay, and among them six molecules exhibited \(>\)50 % inhibition at \(25\,\upmu \hbox {M}\). These molecules also exhibited good growth inhibition against TPBC and TNBC cancer types. Among them, molecules 1 and 17 showed single-digit micromolar \(\hbox {GI}_{50}\) values against MCF-7 and MDA-MB-231 cell lines.









Similar content being viewed by others
References
Oskarsson T (2013) Extracellular matrix components in breast cancer progression and metastasis. Breast 2:S66–72. doi:10.1016/j.breast.2013.07.012
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) The extracellular matrix of animals, 4th edn. Molecular biology of the cell,. Garland Science, New York
Sakkiah S, Thangapandian S, Lee KW (2012) Ligand-based virtual screening and molecular docking studies to identify the critical chemical features of potent cathepsin D inhibitors. Chem Biol Drug Des 80:64–80. doi:10.1111/j.1747-0285.2012.01339.x
Gocheva V, Joyce JA (2007) Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle 6:60–64. doi:10.4161/cc.6.1.3669
Dian D, Vrekoussis T, Shabani N, Mylonas I, Kuhn C, Schindlbeck C, Navrozoglou I, Friese K, Makrigiannakis A, Jeschke U (2012) Expression of cathepsin-D in primary breast cancer and corresponding local recurrence or metastasis: an immunohistochemical study. Anticancer Res 32:901–905
Jacobson-Raber G, Lazarev I, Novack V, Mermershtein W, Baumfeld Y, Geffen DB, Sion-Vardy N, Ariad S (2011) The prognostic importance of cathepsin D and E-cadherin in early breast cancer: a single-institution experience. Oncol Lett 2:1183–1190. doi:10.3892/ol.2011.393
Baldwin ET, Bhat TN, Gulnik S, Hosur MV, Sowder RC, Cachau RE, Collins J, Silva AM, Erickson JW (1993) Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design. Proc Natl Acad Sci USA 90:6796–6800. doi:10.1073/pnas.90.14.6796
Wang F, Samudio I, Safe S (2001) Transcriptional activation of cathepsin D gene expression by 17beta-estradiol: mechanism of aryl hydrocarbon receptor-mediated inhibition. Mol Cell Endocrinol 172:91–103. doi:10.1016/S0303-7207(00)00379-8
Abbott DE, Margaryan NV, Jeruss JS, Khan S, Kaklamani V, Winchester DJ, Hansen N, Rademaker A, Khalkhali-Ellis Z, Hendrix MJ (2010) Reevaluating cathepsin D as a biomarker for breast cancer: serum activity levels versus histopathology. Cancer Biol Ther 9:23–30. doi:10.4161/cbt.9.1.10378
Benes P, Vetvicka V, Fusek M (2008) Cathepsin D-many functions of one aspartic protease. Crit Rev Oncol Hemat 68:12–28. doi:10.1016/j.critrevonc.2008.02.008
Laurent-Matha V, Farnoud MR, Lucas A, Rougeot C, Garcia M, Rochefort H (1998) Endocytosis of pro-cathepsin D into breast cancer cells is mostly independent of mannose-6-phosphate receptors. J Cell Sci 111:2539–2549
Gieselmann V, Hasilik A, Von Figura K (1985) Processing of human cathepsin D in lysosomes in vitro. J Biol Chem 260:3215–3220. http://www.jbc.org/content/260/5/3215.full.pdf
Ahmed W, Jabeen U, Khaliq S (2014) New inhibitors of proteolytic enzymes cathepsin D and plasmepsin II. Pak J Biochem Mol Biol 47:129–132
Fehrenbacher N, Jäättelä M (2005) Lysosomes as targets for cancer therapy. Cancer Res 65:2993–2995. doi:10.1158/0008-5472.CAN-05-0476
Huang L, Liu Z, Chen S, Liu Y, Shao Z (2013) A prognostic model for triple-negative breast cancer patients based on node status, cathepsin-D and Ki-67 index. PLoS One 8:e83081. doi:10.1371/journal.pone.0083081
Wolf M, Clark-Lewis I, Buri C, Langen H, Lis M, Mazzucchelli L (2003) Cathepsin D specifically cleaves the chemokines macrophage inflammatory protein-1 alpha, macrophage inflammatory protein-1 beta, and SLC that are expressed in human breast cancer. Am J Pathol 162:1183–1190. doi:10.1016/S0002-9440(10)63914-4
Khalkhali-Ellis Z, Goossens W, Margaryan NV, Hendrix MJ (2014) Cleavage of Histone 3 by cathepsin D in the involuting mammary gland. PLoS One 9:e103230. doi:10.1371/journal.pone.0103230
Lin T, Williams HR (1979) Inhibition of cathepsin D by synthetic oligopeptides. J Biol Chem 254:11875–11883
Ahmed W, Khan IA, Arshad MN, Siddiqui WA, Haleem MA, Azim MK (2013) Identification of sulfamoylbenzamide derivatives as selective cathepsin D inhibitors. Pak J Pharm Sci 26:687–690
Laurent-Matha V, Derocq D, Prébois C, Katunuma N, Liaudet-Coopman E (2006) Processing of human cathepsin D is independent of its catalytic function and auto-activation: involvement of cathepsins L and B. J Biochem 139:363–371. doi:10.1093/jb/mvj037
Fusek M, Vetvicka V (2005) Dual role of cathepsin D: ligand and protease. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 149:43–50
Grädler U, Czodrowski P, Tsaklakidis C, Klein M, Werkmann D, Lindemann S, Maskos K, Leuthner B (2014) Structure-based optimization of non-peptidic cathepsin D inhibitors. Bioorg Med Chem Lett 24:4141–4150. doi:10.1016/j.bmcl.2014.07.054
Schrodinger, LLC, New York. http://www.schrodinger.com/
McConnell RM, Green AW, Trana CJ, McConnell MS, Lindley JF, Sayyar K, Godwin WE, Hatfield SE (2006) New cathepsin d inhibitors with hydroxyethylamine isosteres: preparation and characterization. Med Chem 2:27–38. doi:10.2174/157340606775197705
Sakkiah S, Meganathan C, Sohn YS, Namadevan S, Lee KW (2012) Identification of important chemical features of \(11\beta \)-hydroxysteroid dehydrogenase type1 inhibitors: application of ligand based virtual screening and density functional theory. Int J Mol Sci 13:5138–5162. doi:10.3390/ijms13045138
Fukuda ME, Iwadate Y, Machida T, Hiwasa T, Nimura Y, Nagai Y, Takiguchi M, Tanzawa H, Yamaura A, Seki N (2005) Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res 65:5190–5194. doi:10.1158/0008-5472.CAN-04-4134
Matarrese P, Ascione B, Ciarlo L, Vona R, Leonetti C, Scarsella M, Mileo AM, Catricala C, Paggi MG, Malomi W et al (2010) Cathepsin B inhibition interferes with metastatic potential of human melanoma: an in vitro and in vivo study. Mol Cancer 9:207–220. doi:10.1186/1476-4598-9-207
Carew JS, Espitia CM, Esquivel JA, Mahalingam D, Kelly KR, Reddy G, Giles FJ, Nawrocki ST (2011) Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem 8:6602–6613. doi:10.1074/jbc.M110.151324
Montcourrier P, Mangeat PH, Salazar G, Morisset M, Sahuquet A, Rochefort H (1990) Cathepsin D in breast cancer cells can digest extracellular matrix in large acidic vesicles. Cancer Res 50:6045–6054
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the \(2^{-\Delta \Delta {\rm c}}\text{ T }\) Method. Methods 25:402–408
Knopfova L, Benes P, Pekarcikova L, Hermanova M, Masarik M, Pernicova Z, Soucek K, Smarda J (2012) c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis. Mol Cancer 11:15. doi:10.1186/1476-4598-11-15
Dali B, Keita M, Megnassan E, Frecer V, Miertus S (2012) Insight into selectivity of peptidomimetic inhibitors with modified statine core for plasmepsin II of Plasmodium falciparum over human cathepsin D. Chem Biol Drug Des 79:411–430. doi:10.1111/j.1747-0285.2011.01276.x
Nakayama S, Torikoshi Y, Takahashi T, Yoshida T, Sudo T, Matsushima T, Kawasaki Y, Katayama A, Gohda K, Hortobagyi GN, Noguchi S, Sakai T, Ishihara H, Ueno NT et al (2009) Prediction of paclitaxel sensitivity by CDK1 and CDK2 activity in human breast cancer cells. Breast Cancer Res 11:1–10. doi:10.1186/bcr2231
Acknowledgments
The authors gratefully acknowledge the Birla Institute of Technology and Sciences, Hyderabad for infrastructural facilities and DST for the INSPIRE fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Anantaraju, H.S., Battu, M.B., Viswanadha, S. et al. Cathepsin D inhibitors as potential therapeutics for breast cancer treatment: Molecular docking and bioevaluation against triple-negative and triple-positive breast cancers. Mol Divers 20, 521–535 (2016). https://doi.org/10.1007/s11030-015-9645-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-015-9645-8