Abstract
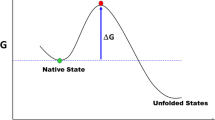
An accurate estimation of the Protein Space size, in light of the factors that govern it, is a long-standing problem and of paramount importance in evolutionary biology, since it determines the nature of protein evolvability. A simple analysis will enable us to, firstly, reduce an unrealistic Protein Space size of ~ 10130 sequences, for a 100-residues polypeptide chain, to ~ 109 functional proteins and, secondly, estimate a robust average-mutation rate per amino acid (ξ ~ 1.23) and infer from it, in light of the protein marginal stability, that only a fraction of the sequence will be available at any one time for a functional protein to evolve. Although this result does not solve the Protein Space vastness problem frames it in a more rational one and illustrates the impact of the marginal stability on protein evolvability.
Similar content being viewed by others
References
Maynard Smith J (1970) Natural selection and the concept of a protein space. Nature 225:563–564
Mandecki W (1998) The game of chess and searches in protein sequence space. Trends Biotechnol 16:200–202
Dryden DTF, Thomson AR, White JH (2008) How much of protein sequence space has been explored by life on Earth? J R Soc Interface 5:953–956
Romero PA, Arnold FH (2009) Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol 10:866–876
Ivankov DN (2017) Exact correspondence between walk in nucleotide and protein sequence spaces. PLoS ONE 12(8):e0182525
Levinthal C (1968) Are there pathways for protein folding? J de Chim Phys 65(1):44–45
Zwanzig R, Szabo A, Bagchi B (1992) Levinthal’s paradox. Proc Natl Acad Sci USA 89:20–22
Finkelstein AV, Garbuzynskiy SO (2013) Levinthal’s question answered…again? J Biomol Struct Dyn 31(9):1013–1015
Wagner A (2005) Robustness, evolvability, and neutrality. FEBS Lett 579:1772–1778
DePristo M, Weinreich D, Hartl D (2005) Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet 6:678–687
Bloom JD, Labthavikul ST, Otey CR, Arnold FH (2006) Protein stability promotes evolvability. Proc Natl Acad Sci USA 103:5869–5874
Zeldovich KB, Chen P, Shakhnovich EI (2007) Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA 104:16152–16157
Tokuriki N, Tawfik DS (2009) Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19:596–604
Tokuriki N, Stricher F, Serrano L, Tawfik DS (2008) How protein stability and new functions trade off. PLoS Comput Biol 4(2):e1000002
Kurahashi R, Sano S, Takano K (2018) Protein evolution is potentially governed by protein stability: directed evolution of an esterase from the hyperthermophilic archaeon sulfolobus tokodaii. J Mol Evol 86(5):283–292. https://doi.org/10.1007/s00239-018-9843-y
Otwinowski J (2018) Biophysical Inference of epistasis and the effects of mutations on protein stability and function. Mol Biol Evol 35(10):2345–2354
Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA (2012) Epistasis as the primary factor in molecular evolution. Nature 490:535–538
Starr TN, Thornton JW (2016) Epistasis in protein evolution. Protein Sci 25:1204–1218
Miton CM, Chen JZ, Ost K, Anderson DW, Tokuriki N (2020) Statistical analysis of mutational epistasis to reveal intramolecular interaction networks in proteins. Methods Enzymol 643:243–280. https://doi.org/10.1016/bs.mie.2020.07.012
Koehl P, Levitt M (2002) Protein topology and stability define the space of allowed sequences. Proc Natl Acad Sci USA 99(3):1280–1285
Privalov PL, Tsalkova TN (1979) Micro- and macro-stabilities of globular proteins. Nature 280:694–696
Hormoz S (2013) Amino acid composition of proteins reduces deleterious impact of mutations. Sci Rep 3:1–10
Vila JA (2019) Forecasting the upper bound free energy difference between protein native-like structures. Phys A 533:122053
Martin OA, Vila JA (2020) The marginal stability of proteins: how the jiggling and wiggling of atoms is connected to neutral evolution. J Mol Evol 88:424–426
Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–323
Cecchini M, Krivov SV, Spichty M, Karplus M (2009) Calculation of free-energy differences by confinement simulations. Application to peptide conformers. J Phys Chem B 113:9728–9740
Spichty M, Cecchini M, Karplus M (2010) Conformational free-energy difference of a miniprotein from nonequilibrium simulations. J Phys Chem Lett 1(13):1922–1926
Vila JA (2020a) Metamorphic proteins in light of Anfinsen’s Dogma. J Phys Chem Lett 11(13):4998–4999
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Gillespie JH (1994) The causes of molecular evolution. Oxford University Press, Incorporated, Oxford
Lipman DJ, Wilbur WJ (1991) Modelling neutral and selective evolution of protein folding. Proc Royal Soc Lond B 245:7–11
Bornberg-Bauer E (1997) How are model protein structures distributed in sequence space? Biophys J 73(5):2393–2403
Wolynes P (1997) As simple as can be? Nat Struct Mol Biol 4:871–874
Perutz MF (1983) Species adaptation in a protein molecule. Mol Biol Evol 1(1):1–28
Wilson AE, Kosater WM, Liberles DA (2020) Evolutionary processes and biophysical mechanisms: revisiting why evolved proteins are marginally stable. J Mol Evol 88:415–417
Margoliash E, Smith EL (1965) Structural and functional aspects of cytochrome c in relation to evolution. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins: a symposium. Academic Press, New York, London, pp 221–242
Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV et al (2016) Local fitness landscape of the green fluorescent protein. Nature 533:397–401
Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626
Otha T (2006) Slightly deleterious mutant substitutions in evolution. Nature 246:96–97
Schopf JW (2006) The first billion years: When did life emerge? Elements 2:229–233
Koonin E, Wolf Y, Karev G (2002) The structure of the protein universe and genome evolution. Nature 420:218–223. https://doi.org/10.1038/nature01256
Sweetlove L (2011) Number of species on Earth tagged at 8.7 million. Nature. https://doi.org/10.1038/news.2011.498
Vila JA (2020b) Harold A. Scheraga Legatum. Protein J. https://doi.org/10.1007/s10930-020-09917-w
Acknowledgements
I would like to thank to Laura Mascotti, Walter Lapadula and Maximiliano Juri Ayub for reading the manuscript and making valuable comments and suggestions. The author acknowledges financial support from the IMASL-CONICET (PIP-0087) and ANPCyT (Grant Nos. PICT-0767; and PICT-2212), Argentina.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vila, J.A. About the Protein Space Vastness. Protein J 39, 472–475 (2020). https://doi.org/10.1007/s10930-020-09939-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09939-4