Abstract
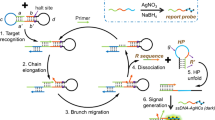
MicroRNAs (miRNA) are a novel class of small noncoding RNAs with roles in RNA silencing and post transcriptional regulation of gene expression. Due to their roles, miRNA can be considered as new biomarkers for prognosis of diseases such as Multiple sclerosis (MS). Herein, we report a miRNA nanobiosensor based on nucleic acid hybridization chain reaction and highly fluorescent DNA hosted silver nanoclusters (NC). In our method, two types of hairpin oligonucleotide probes, MB1 and MB2, were employed as hybridization chain reaction (HCR) monomers, where MB1 acted as a template for in situ synthesis of fluorescent Ag NC. These monomers were stable in solution but they triggered a cascade of hybridization events once miR-145 (a biomarker of MS in blood) was added to the solution. The process yielded nicked double stranded DNA. The nanobiosensor showed great sensitivity for the detection of target microRNA and excellent limit of detection of about 0.1 nM with high specificity to differentiate sharply between complementary, mismatch, and non-complementary target miRNAs. Alongside the outstanding sensitivity and selectivity, the nanobiosensor exhibited great reproducibility, stability and a decent response in real sample analysis with blood plasma. In conclusion, this simple and highly responsive nanobiosensor can clinically be used for the early detection of MS by direct detection of the plasma miR-145 in real clinical samples, without a need for sample preparation, RNA extraction and/or amplification.






Similar content being viewed by others
References
Milo R, Kahana E (2010) Multiple sclerosis: Geoepidemiology, genetics and the environment. Autoimmun Rev 9:387–394
Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y et al (2014) Expression, regulation and function of MicroRNAs in multiple sclerosis. Int J Med Sci 11:810–818
Siege SR, MacKenzie J, Chaplin G, Jablonski NG, Griffiths L (2012) Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep 39:6219–6225
Dyment DA, Ebers GC, Dessa Sadovnick A (2004) Genetics of multiple sclerosis. The Lancet Neurology 3(2):104–110
Sospedra M, Martin R (2005) Immunology of multiple sclerosis. Annu Rev Immun 23:683–747
de Jr Omar F, Moore CS, Kennedy TE, Antel JP, Bar-Or A, Dhaunchak AS (2013) MicroRNA dysregulation in multiple sclerosis. Front Genet 3:1–6
Fenoglio C, Cantoni C, De Riz M, Ridolfi E, Cortini F, Serpente M et al (2011) Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci Lett 504:9–12
Martinelli-Boneschi F, Fenoglio C, Brambilla P, Sorosina M, Giacalone G, Esposito F et al (2012) MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci Lett 508:4–8
Fenoglio C, Ridolfi E, Galimberti D, Scarpini E (2012) MicroRNAs as active players in the pathogenesis of multiple sclerosis. Int J Mol Sci 13:13227–13239
Schöler N, Langer C, Döhner H, Buske C, Kuchenbauer F (2010) Serum microRNAs as a novel class of biomarkers: a comprehensive review of the literature. Exp Hemat 38:1126–1130
Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M et al (2009) Multiple sclerosis: MicroRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One 4:e7440
Guerau-de-Arellano M, Alder H, Ozer HG, Lovett-Racke A, Racke MK (2012) miRNA profiling for biomarker discovery in multiple sclerosis: from microarray to deep sequencing. J Neuroimmunol 248:32–39
Thamilarasan M, Koczan D, Hecker M, Paap B, Zettl UK (2012) MicroRNAs in multiple sclerosis and experimental autoimmune encephalomyelitis. Autoimmun Rev 11:174–179
Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50(4):298–301
Li W, Ruan K (2009) MicroRNA detection by microarray. Anal Bioanal Chem 394:1117–1124
Zhang P, Wu X, Chai Y, Yuan R (2014) An electrochemiluminescent microRNA biosensor based on hybridization chain reaction coupled with hemin as the signal enhancer. Analyst 139(11):2748–2753
Azimzadeh M, Rahaie M, Nasirizadeh N, Ashtari K, Naderi-Manesh H (2016) An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens Bioelectron 77:99–106
Samanta A, Banerjee S, Liu Y (2015) DNA nanotechnology for nanophotonic applications. Nano 7:2210–2220
Pinheiro AV, Han D, Shih WM, Yan H (2011) Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol 6:763–772
Qiu X, Wang P, Cao Z (2014) Hybridization chain reaction modulated DNA-hosted silver nanoclusters for fluorescent identification of single nucleotide polymorphisms in the let-7miRNAfamily. Biosens Bioelectron 60:351–357
Volkov I, Sych T, Serdobintsev P, Reveguk Z, Kononov A (2016) Fluorescence saturation spectroscopy in probing electronically excited states of silver nanoclusters. J Luminescence 172:175–179
Zhao H, Wang L, Zhu J, Wei H, Jiang W (2015) Label-free nucleicacids detection based on DNA templated silver nanoclusters fluorescent probe. Talanta 138:163–168
Dong H, Hao K, Tian Y, Jin S, Lu H, Zhou S-F, Zhang X (2014) Label-free and ultrasensitive microRNA detection based on novel molecular beacon binding readout and target recycling amplification. Biosens Bioelectron 53:377–383
Guo W, Yuan J, Dong Q, Wang E (2010) Highly sequence-dependent formation of fluorescent silver nanoclusters in hybridized DNA duplexes for single nucleotide mutation identification. J Am Chem Soc 132:932–934
Choi HMT, Beck VA, Pierce NA (2014) Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8:4284–4294
Wang W, Zhan L, Du YQ, Leng F, Chang Y, Gao MX, Huang CZ (2013) Label-free DNA detection on the basis of fluorescence resonance energy transfer from oligonucleotide-templated silver nanoclusters to multi-walled carbon nanotubes. Anal Methods 5:5555–5559
Rahaie M, Ghai R, Babic B, Dimitrov K (2009) Synthesis and Characterization of DNA-based micro- and Nanodumbbell structures. J Bionanosci 3:73–79
Obliosca JM, Babin MC, Liu C, Liu YL, Chen Y, Batson RA, Ganguly M, Petty JT, Yeh HC (2014) A complementary palette of NanoCluster beacons. ACS Nano 8:10150–10160
O’Neill PR, Velazquez LR, Dunn DG, Gwinn EG, Fygenson DK (2009) Hairpins with poly-C loops stabilize four types of fluorescent Agn:DNA. J Phys Chem C 113:4229–4233
Gwinn EG, O'Neill P, Guerrero AJ, Bouwmeester D, Fygenson DK (2008) Sequence-dependent fluorescence of DNA-hosted silver nanoclusters. Adv Mat 20:279–283
Yeh H-C, Sharma J, Han JJ, Martinez JS, Werner JH (2010) A DNA-silver nanocluster probe that fluoresces upon hybridization. Nano Lett 10:3106–3311
Zhou Y, Yin H, Li J, Li B, Li X, Ai S, Zhang X (2016) Electrochemical biosensor for microRNA detection based on poly (U) polymerase mediated isothermal signal amplification. Biosens Bioelectron 79:79–85
Liu Y-Q, Zhang M, Yin B-C, Ye B-C (2012) Attomolar ultrasensitive MicroRNA detection by DNA-Scaffolded silver-nanocluster probe based on isothermal amplification. Anal Chem 84(12):5165–5169
Acknowledgments
We would to thank the University of Tehran for providing financial and instrumental supports in this work. This work was supported by University of Tehran (Grant Number, 104417). Authors also thank Iman Daryaei from University of Arizona, USA, for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mansourian, N., Rahaie, M. & Hosseini, M. A Nanobiosensor Based on Fluorescent DNA-Hosted Silver Nanocluster and HCR Amplification for Detection of MicroRNA Involved in Progression of Multiple Sclerosis. J Fluoresc 27, 1679–1685 (2017). https://doi.org/10.1007/s10895-017-2105-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-017-2105-3