Abstract
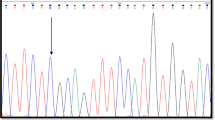
Familial Hemophagocytic Lymphohistiocytosis type 2 (FHL2) results from mutations in PRF1. We described two unrelated individuals who presented with FHL, in whom severely impaired NK cytotoxicity and decrease perforin expression was observed. DNA sequencing of PRF1 demonstrated that both were not only heterozygous for the p.54R > C/91A > V haplotype but also presented with the novel variant p.47G > V at the perforin protein. Perforin mRNA was found to be increased in a individual with that genotype. A carrier of the novel variant also demonstrated altered perforin mRNA and protein expression. Phylogenetic analysis and multiple alignments with perforin orthologous demonstrated a high level of conservation at Gly47. PolyPhen-2 and PROVEAN predicted p.47G > V to be “probably damaging” and “deleterious”, respectively. A thermodynamic analysis showed that this variant was highly stabilizing, decreasing the protein internal energy. The ab initio perforin molecular modeling indicated that Gly47 is buried inside the hydrophobic core of the MACPF domain, which is crucial for the lytic pore formation and protein oligomerization. After the in silico induction of the p.47G > V mutation, Val47 increased the interactions with the surrounding amino acids due to its size and physical properties, avoiding a proper conformational change of the domain. To our knowledge, this is the first description supporting that p.47G > V is a pathogenic variant that in conjunction with p.54R > C/91A > V might result in the clinical phenotype of FHL2.





Similar content being viewed by others
References
Goransdotter EK, Fadeel B, Nilsson-Ardnor S, Söderhäll C, Samuelsson A, Janka G, et al. Spectrum of perforin gene mutations in familial hemophagocytic lymphohistiocytosis. Am J Hum Genet. 2001;68(3):590–7.
Trizzino A, ZurStadt U, Ueda I, Risma K, Janka G, Ishii E, et al. Genotype–phenotype study of familial haemophagocytic lymphohistiocytosis due to perforin mutations. J Med Genet. 2008;45(1):15–21.
Kogawa K, Lee SM, Villanueva J, Marmer D, Sumegi J, Filipovich AH. Perforin expression in cytotoxic lymphocytes from patients with hemophagocytic lymphohistiocytosis and their family members. Blood. 2002;99(1):61–6.
Feldmann J, Le Deist F, Ouachee-Chardin M, Certain S, Alexander S, Quartier P, et al. Functional consequences of perforin gene mutations in 22 patients with familial haemophagocytic lymphohistiocytosis. Br J Haematol. 2002;117(4):965–72.
Janka GE. Familial and acquired hemophagocytic lymphohistiocytosis. Annu Rev Med. 2012;63:233–46.
Voskoboinik I, Smyth MJ, Trapani JA. Perforin- mediated target-cell death and immune homeostasis. Nat Rev Immunol. 2006;6:940–52.
Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew PA, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286(5446):1957–9.
Baran K, Dunstone M, Chia J, et al. The Molecular basis for Perforin Oligomerization and Transmembrane Pore Assembly. Immunity. 2009;30(5):684–95.
Law RH, Lukoyanova N, Voskoboinik I, et al. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468(7322):447–51.
Zhang K, Jordan MB, Marsh RA, et al. Hypomorphic mutations in PRF1, MUNC13-4, and STXBP2 are associated with adult-onset familial HLH. Blood. 2011;118(22):5794–8.
Zhang K, Chandrakasan S, Chapman H, et al. Synergistic defects of different molecules in the cytotoxic pathway lead to clinical familial hemophagocytic lymphohistiocytosis. Blood. 2014;124(8):1331–4.
Mhatre S, Madkaikar M, Desai M, Ghosh K. Spectrum of perforin gene mutations in familialhemophagocytic lymphohistiocytosis (FHL) patients in India. Blood Cells Mol Dis. 2015;54(3):250-7.
Risma KA, Frayer RW, Filipovich AH, et al. Aberrant maturation of mutant perforin underlies the clinical diversity of hemophagocytic lymphohistiocytosis. J Clin Invest. 2006;116(1):182–92.
Peitsch MC, Amiguet P, Guy R, Brunner J, Maizel Jr JV, Tschopp J. Localization and molecular modelling of the membrane-inserted domain of the ninth component of human complement and perforin. Mol Immunol. 1990;27(7):589–602.
Sánchez IP, Leal-Esteban LC, Álvarez-Álvarez JA, et al. Analyses of the PRF1 gene in individuals with hemophagocytic lymphohystiocytosis reveal the common haplotype R54C/A91V in Colombian unrelated families associated with late onset disease. J Clin Immunol. 2012;32(4):670–80.
Henter JI, Horne A, Aricó M, Egeler RM, Filipovich AH, Imashuku S, et al. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48(2):124–31.
Li S, Ma D, Zhang L, et al. High levels of FCcR3A and PRF1 expression in peripheral blood mononuclear cells from patients with primary biliary cirrhosis. Dig Dis Sci. 2013;58:458–64.
Chia J, Thia K, Brennan AJ, Little M, Williams B, Lopez JA, et al. Fatal immune dysregulation due to a gain of glycosylation mutation in lymphocyte perforin. Blood. 2012;119(7):1713–6.
National Center for Biotechnology Information, PRF1 gene reference sequence information (http://www.ncbi.nlm.nih.gov/gene/5551 )
Hall, T. BioEdit: Biological sequence alignment software. Ibis Biosciences. Version 7.2.3. http://www.mbio.ncsu.edu/BioEdit/bioedit.html
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. http://genetics.bwh.harvard.edu/pph2/.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid aubstitutions and indels. PLoS ONE. 2012;7(10):e46688. PROVEAN (Protein Variation Effect Analyzer). (http://provean.jcvi.org/seq_submit.php).
Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011;12:151. http://babylone.ulb.ac.be/PoPV2a/index.php.
Worth CL, Preissner R, Blundell TL. SDM--a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39 (Web Server issue):W215-22. (http://mordred.bioc.cam.ac.uk/sdm/sdm.php)
UniProt Consortium 2013, UniProtKB Protein Knowledgebase. http://www.uniprot.org/
Larkin MA, Blackshields G, Brown NP, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23(21):2947–8. http://www.clustal.org.
Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. http://www.megasoftware.net.
Berman HM, Battistuz T, Bhat TN, et al. The protein data bank. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 6 No 1):899–907. http://www .pdb.org/.
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. http://zhanglab.ccmb.med.umich.edu/I-TASSER/.
The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. http://www.cgl.ucsf.edu/chimera.
Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–23. http://spdbv.vital-it.ch/.
Julenius K. NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology. 2007;17(8):868–76. http://www.cbs.dtu.dk/services/NetCGlyc/.
Gupta R, Jung E, Brunak S. Prediction of N-glycosylation sites in human proteins. In preparation, 2004. http://www.cbs.dtu.dk/services/NetNGlyc/
Steentoft C, Vakhrushev SY, Joshi HJ, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–88. http://www.cbs.dtu.dk/services/NetOGlyc/.
Online Mendelian Inheritance in Man, OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: {170280}: {12/06/2013}. http://omim.org/
An O, Gursoy A, Gurgey A, Keskin O. Structural and functional analysis of perforin mutations in association with clinical data of familial hemophagocyticlymphohistiocytosis type 2 (FHL2) patients. Protein Sci. 2013;22(6):823–39.
Ramachandran S, Kota P, Ding F, Dokholyan NV. Automated minimization of steric clashes in protein structures. Proteins. 2011;79(1):261–70.
Clementi R, Emmi L, Maccario R, Liotta F, Moretta L, Danesino C, et al. Adult onset and atypical presentation of hemophagocytic lymphohistiocytosis in siblings carrying PRF1 mutations. Blood. 2002;100(6):2266–7.
Ueda I, Kurokawa Y, Koike K, Ito S, Sakata A, Matsumora T, et al. Late-onset cases of familial hemophagocytic lymphohistiocytosis with missense perforin gene mutations. Am J Hematol. 2007;82(6):427–32.
Kim N, Kim M, Yun S, et al. MicroRNA-150 regulates the cytotoxicity of natural killers by targeting perforin-1. J Allergy Clin Immunol. 2014;134(1):195–203.
Marcenaro S, Gallo F, Martini S, et al. Analysis of natural killer-cell function in familial hemophagocytic lymphohistiocytosis (FHL): defective CD107a surface expression heralds Munc13-4 defect and discriminates between genetic subtypes of the disease. Blood. 2006;108(7):2316–23.
Bryceson YT, Pende D, Maul-Pavicic A, et al. A prospective evaluation of degranulation assays in the rapid diagnosis of familial hemophagocytic syndromes. Blood. 2012;119(12):2754–63.
Acknowledgments
We are grateful to the patients’ families and healthy control volunteers involved in this study for their participation and cooperation. JAP acknowledges the grants from Vicerrectoría de Investigación (CIQF-177) and Estrategia de Sostenibilidad 2013–2014, Universidad de Antioquia. We thank Javier Rendón for his medical advice. This study was supported by the Colombian Institute for the development of Science and Technology, COLCIENCIAS (Grant #111556934426) and Estrategia de Sostenibilidad 2011–2012, Vicerrectoría de Investigaciones-Universidad de Antioquia, Medellin- Colombia.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Romero, C.A.P., Sánchez, I.P., Gutierrez-Hincapié, S. et al. A Novel Pathogenic Variant in PRF1 Associated with Hemophagocytic Lymphohistiocytosis. J Clin Immunol 35, 501–511 (2015). https://doi.org/10.1007/s10875-015-0169-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-015-0169-x