Abstract
The crystal structures of (1-phenyl-1H-1,2,3-triazol-4-yl)methyl benzoate, 1a, (2-(4-fluorophenyl)-2H-1,2,3-triazole-4-yl)methanol, 2a, and 2-phenyl-2H-1,2,3-triazol-4-carbaldehyde, 2b, are reported. Compounds 1a and 2a were recently reported to exhibit mild α-glycosidase inhibition activity, while compound 2b exhibited a much greater activity. Only small dihedral angles 6.52(4), 14.02(10) and 2.44(7)° are present between the triazolyl ring and the attached aryl rings in 1a, 2a and 2b, respectively. The relatively flat compounds 2a and 2b contrast with compound 1a, which is “V” shaped, with a dihedral angle between the near planar phenyltriazolyl-CH2 and phenyl-CO2CH2 moieties of 88.11(4)°. The intermolecular interactions in 1a are C–H···X (X = N or π(triazole) and π(triazole) ···π(phenyl): two different chains are formed, from (i) combinations of the C–H···N hydrogen bonds and (ii) combinations of the C–H···π and π···π interactions.. The intermolecular interactions in 2a are C–H···O and C–F···π(phenyl): the C–H···O interactions generate a sheet of molecules, containing a network of rings.·Classical O–H···O hydrogen bonds, and weaker C–H···π(triazolyl) and π(phenyl)···π(triazolyl) interactions are present in 2b: all three interactions together generate a chevron-type arrangement. Compound 1a crystallizes in the monoclinic space group P21 with a = 4.5661(5), b = 10.5573(14), c = 13.9694(19) Å, β = 90.594(6)° and Z = 2. Compound 2a crystallizes in the monoclinic space group P21 with a = 3.7175(7), b = 10.428(2), c = 10.689(3) Å, β = 90.521(6)° and Z = 2. Compound 2b crystallizes in the monoclinic space group P21/c with a = 11.4130(5), b = 4.80280(10), c = 15.5916(11) Å, β = 103.373(7)° and Z = 4.
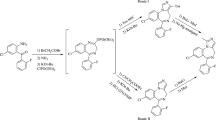
Graphical Abstract
The relatively flat compounds, (2-(4-fluorophenyl)-2H-1,2,3-triazole-4-yl)methanol and 2-phenyl-2H-1,2,3-triazol-4-carbaldehyde, contrast with compound (1-phenyl-1H-1,2,3-triazol-4-yl)methyl benzoate, which is “V” shaped, with a dihedral angle between the near planar phenyltriazolyl-CH2 and phenyl-CO2CH2 moieties of 88.11(4) o.

Similar content being viewed by others
Introduction
1,2,3-Triazole exists in two tautomeric forms, namely 1H-1,2,3-triazole and 2H-1,2,3-triazole, see Fig. 1. Derivatives of both forms have attracted much attention [1–3], in particular for their biological activities, which include as antiviral [4–6], antimalarial [7], antitubercular [8–10], antifungal [11, 12] anti-HIV [13], β-lactamase inhibition [14], anti-epileptic [15], anti-HSV [16], anti-inflammatory [17], antimicrobial [18, 19] and α-glycosidase inhibition agents [20–23]. Patents lodged in the period 2008–2011 for 1H-1,2,3-triazole and 2H-1,2,3-triazole derivatives have been included in a general survey for all triazolyl compounds [24].
A recent α-glycosidase inhibition study [23] involved a number of different 1-phenyl-1H- and 2-phenyl-2H-1,2,3-triazol derivatives. The crystal structures of three of the compounds from that study [20] have been determined, namely, (1-phenyl-1H-1,2,3-triazol-4-yl)methyl benzoate, 1a, (2-(4-fluorophenyl)-2H-1,2,3-triazole-4-yl)methanol, 2a and 2-phenyl-2H-1,2,3-triazole-4-carbaldehyde, 2b, see Table 1. Compounds, 1a and 2a, exhibited little activity, while compound 2b, exhibited a greater inhibition, as did all the 2-aryl-2H-1,2,3-triazole-4-carbaldehyde derivatives. It was suggested that the aldehydes act upon both yeast maltase and PPA, with the aldehyde groups reacting with amine groups in the enzyme polypeptide chain to form Schiff bases.
We now wish to report our structural findings.
Results and Discussion
The compounds were prepared as previously reported, see Scheme 1 [20].
Molecular Confirmations
Compounds 2a and 2b crystallize in the monoclinic space group, P21 with Z = 2, while compound 2b crystallizes in the monoclinic space group, P21/c with Z = 4. The asymmetric unit in each case consists of a single molecule, as illustrated in Fig. 2. Selected bond lengths and angles are listed in Table 2. The bond lengths and angles associated with the 1,2,3-triazole rings are in the regions normally found for 1-aryl-1H-1,2,3,triazoles [see for example 25–28] and for 2-aryl-2H-1,2,3,triazoles [see for example Refs. 28–33].
In each of 1a, 2a and 2b, the triazolyl ring is essentially planar. The dihedral angles between the triazolyl and aryl rings are listed in Table 3.
A large range of dihedral angles have been reported for both 1-aryl-1H-1,2,3-triazole and 2-aryl-2H-1,2,3-triazole compounds, for example the angles are 0.34(17) and 87.1(2)°, respectively, in 4-difluoromethyl-1-(4-methylphenyl)-1H-1,2,3-triazole, 1b, [34] and in one independent molecule of 1-[5-methyl-1-(4-nitrophenyl)-1H-1,2,3-triazol-4-yl]ethanone, 1c [35], see Table 1. The carbon atoms of the methylene units in 2b and 1a are essentially co-planar with the attached triazolyl group. Compound 1a has a “V” shape with the angle between the near planar phenyltriazolyl-CH2 and phenyl-CO2CH2 moieties of 88.11(4)°.
Crystal Structures
Compound 1a
The intermolecular interactions in 1a are all weak, being C–H···N hydrogen bonds, C–H···π and π–π interactions. As illustrated in Fig. 3a, chains of molecules are propagated in the direction of the b axis, from combinations of C5–H5···N3, C5–H5···N2 and C8–H8···N3 hydrogen bonds, see Table 4 for the symmetry operations. Of these hydrogen bonds, the C5–H5···N3 is the most significant. With C5–H5 acting as donor to two acceptors, N2 and N3, \({\text{R}}^{1}_{2}\) (3) rings are generated. The C5–H5···N3, C5–H5···N2, and C8–H8···N3 hydrogen bonds individually generate C4, C4 and C6 chains of molecules.
Compound 1a. a A chain of molecules generated in the direction of the b axis, from combinations of C5–H5···N3, C5–H5···N2 and C8–H8···N3 hydrogen bonds, b a further chain of molecules, propagated in the direction of the a axis, formed from C6–H6A···π(triazole)and weak π(phenyl)···π(triazole) interactions. Table 4 lists the symmetry operations. Intermolecular interactions are drawn as thin dashed lines
Further, chain of molecules, this time propagated in the direction of the a axis, are formed from combinations of C6–H6A···π(triazolyl) and π(phenyl)···π(triazolyl) stacking interactions, see Fig. 3b; Table 4. This combination of interactions provides a chevron-type arrangement. The perpendicular distance between the best planes of combined triazolyl/ phenyl rings between layers is 3.359(3) Å, and with the Cg···Cg separations of 3.738(3) Å, these π···π interactions are important. The packing of the molecules looking down the b axis is shown in Fig. 4.
The structure of a related compound, 1-phenyl-4-(pyridine-3-yl-CO2CH2)-1H-1,2,3-triazole) (1d) [36] has been reported. There are some structural similiarities between 1a and 1d: (i) the molecule of 1d is also “V” shaped with the angle between the planar phenyltriazolyl CH2 and phenyl-CO2CH2 moieties of 83.84°, (ii) the dihedral angle between the triazolyl and phenyl rings is 16.54 (11)°, and (iii) there are similar C–H··· π (triazole) and π(phenyl)···π(triazolyl) interactions [Cg···Cg = 3.895(1) Å]. However other intermolecular interactions are different in 1d, being C–H···N(py), C–H···O(carbonyl) and C–H···π(triazolyl) hydrogen bonds, which generate a different supramolecular array to that of 1a.
Compound 2a
The intermolecular interactions in 2a are C8–H8···O2 and C11–H11···O2 hydrogen bonds and C10–F1··· π(phenyl) interactions. Table 4 lists the symmetry operations and geometric parameters. Combinations of the C8–H8···O2 and C11–H11···O2 hydrogen bonds generate sheets of molecules, composed of \({\text{R}}^{ 4}_{ 3}\) (24) rings, see Fig. 5a. The sheet undulates in the direction of the b axis, as shown in Fig. 4b. Both the C–H···O hydrogen bonds building the sheets are on the weak side. Stacks of molecules are generated from C10–F1···π(phenyl) interactions, see Fig. 5c. The Cg(phenyl)–Cg(triazolyl) distances in adjacent layers within the stacks are 4.2623(17) Å, which suggest any π(phenyl)– π(triazolyl) must be very weak. Figure 6 illustrates the packing of molecules of 2a.
Compound 2a. a An undulating sheet of molecules of 2a, formed from weak C11–H11···O2 and C8–H8···O2 hydrogen bonds, and composed of \({\text{R}}^{ 4}_{ 3}\) (24) rings, b an alternate view of the sheet shown in a, clearly indicating its undulating nature and its alignment along the b axis, c a stack of molecules, with π(phenyl)–π(triazole) stacking interactions, augmented by C10–F1···π(phenyl) interactions. Intermolecular interactions are drawn as thin dashed lines. Table 4 lists the symmetry operations
Compound 2b
Present in 2b are classical O1–H1···O1 hydrogen bonds, and C6–H6B···π(triazolyl) and π(phenyl)···π(triazolyl) interactions. All three interactions together generate a chevron-type arrangement of molecules as illustrated in Fig. 7. The most significant of these interactions are O1–H1···O1 hydrogen bonds, which forms chains of molecules in the direction of the b axis. The packing of the molecules of 2b, looking down the b axis is illustrated in Fig. 8. As in compound 2a, none of the triazolyl nitrogen atoms are involved in any intermolecular interaction in 2b.
Chevron type arrangement of molecules of 2b, generated from strong classical O1–H1···O 1 intermolecular hydrogen bonds and weaker C6–H6B···π(triazole) and π(triazole)···π(phenyl) interactions. Table 4 lists the symmetry operations. Intermolecular interactions are drawn as thin dashed lines
Related Compounds
Comparisons of the structure of 2a can be made with those reported for 1-(4-methylphenyl)-4-OCH-1H-1,2,3-triazole, 1e, [37] and 1-(4-nitrophenyl)-4-trimethylsilyl-1H-1,2,3-triazol-5-carbaldehyde, 1f, [38].
The structure of 1e, which was only briefly discussed in the original article [37], exhibits significant differences with that of 2a. Features of the structure of 1e are (i) a near planar molecule, as shown by the dihedral angle of 7.1° between the triazolyl and the phenyl rings, (ii) C–H(phenyl)···O(=C) and C5–H5···N3 hydrogen bonds forming chains containing \({\text{R}}^{ 2}_{ 2}\) (10) rings propagated in the direction of the a axis, and relatively strong π(triazolyl)···π(triazolyl) and (iii) π(phenyl)···π(phenyl) interactions.in which the Cg···Cg distances and perpendicular distances between planes through the phenyltriazole fragments, in both cases are 3.865(4) and 3.436(4) Å, respectively.
The position of the aldehyde group on C5 of the triazole ring in 1f, results in a much larger dihedral angle between the triazoleyland aryl planes of 62.34(5)°, compared to those in 2a and 1e. Again the carbonyl oxygen is involved in C–H···O hydrogen bonds, this time with a CH unit in the trimethylsilyl group: these C–H···O hydrogen bonds produce chains of molecules.
Comparison of the structure of 2a can be made with the hemihydrate of 1-(3-C6H4)-4-HOCH2-1H-1,2,3-triazole, 1g, [39], 1-(3,5-dimethylphenyl)-4-HOCH2-1H-1,2,3-triazole, 1 h, [40] and 1-(4-biphenyl)-4-HOCH2-1H-1,2,3-triazole. 1i [40] and 1-(4-HO2CC6H4-4-HOCH2-1H-1,2,3-triazole. 1j [41], 1-(2-HO2CC6H4-4-HOCH2-1H-1,2,3-triazole. 1k [41] and 1-(2-HOC6H4-4-HOCH2-1H-1,2,3-triazole. 1l [41].
In both 1-(4-biphenyl)-4-HOCH2-1H-1,2,3-triazole, 1i, and 1-(3,5-dimethylphenyl)-4-HOCH2-1H-1,2,3-triazole, 1h, [40], there are O–H···N3 hydrogen bonds involving the hydroxyl group. However in 1i, these generate chains of molecules, while in 1h centrosymmetric dimers, having \({\text{R}}^{ 2}_{ 2}\) (10) rings, are formed. The dihedral angles between the triazolyl and phenyl rings are 25.29(5) and 23.71(5)° in 1i and 1h, respectively, and thus are much larger than that in 2b [2.44°] and must be a consequence of the crystal packing rather than any steric effect arising from the substituents. Neither 1i nor 1h exhibits π···π stacking interactions.
In the hemihydrate of 1-(3-ClC6H4)-4-HOCH2-1H-1,2,3-triazole, [1g.0.5(H 2 O)] [39], the hydrate plays a significant role in the supramolecular array. The most important intermolecular interactions in [1g.0.5(H 2 O] are Ow–Hw···N3 and Ow–Hw···Ow hydrogen bonds, which generate chains of molecules of water and 1g propagated in the direction of the a-axis. Additional features of the structure are O6H6 twelve-membered rings formed from O–H···O hydrogen bonds involving two hydrate molecules and the hydroxyl groups of two molecules each of the two independent molecules of 1g. Also present in [(1g)2(H2O)] are weak π(phenyl)···π(phenyl) interactions.
As illustrated by [1g.0.5(H 2 O)], the presence of additional donor centres radically changes the involvement of the triazole bound hydroxyl group. This is also very clearly illustrated by the group of compounds, 1j–1l [26], in which the hydroxyl group on the triazole ring preferentially links with the hydroxyl or carboxylic acid substituents on the phenyl ring, leading to the formation of helices and other supramolecular architectures.
Conclusion
There appears to be no consistent intermolecular interaction, nor dihedral angle between the aryl and triazole rings, in either of the two series of aryl-1,2,3-triazoles . The dependence of the supramolecular array on the substituent(s) is clearly apparent.
Experimental
The compounds, 1a, 2a and 2b, were prepared as reported [23]. For the structure determinations, crystals of 1a were obtained from MeOH, 2a from EtOH and 2b from Me2CO solutions.
X-Ray Crystallography
Data for compounds 1a and 2b were obtained at 120(2) K while data for compound 2a were collected at 100(2) K, all with Mo-Kα radiation by means of a Bruker–Nonius Roper CCD camera on kappa-goniostat instrument of the NCS crystallographic service, based at the University of Southampton. Data collection, data reduction and unit cell refinement were achieved with DENZO [41] and COLLECT [42] programs. Correction for absorption was achieved in each program SADABS 2007/2 [43]. The program MERCURY [44] was used in the preparation of the Figures. SHELXL97 [45] and PLATON [46] were used in the calculation of molecular geometry. The structures were solved by direct methods using SHELXS-97 [45] and fully refined by means of the program SHELXL-97 [45]. Difference map provided position for the aldehydic hydrogen atoms of 2b. All other hydrogen atoms were placed in calculated positions. Crystal data and structure refinement details are listed in Table 5.
Supplementary Material
Full details of the crystal structure determinations in cif format are available in the online version, at doi: (to be inserted), and have also been deposited with the Cambridge Crystallographic Data Centre with deposition numbers, 1417606, 1417783 and 1417607, respectively for 1a, 2a and 2b. Copies of these can be obtained free of charge on written application to CCDC, 12 Union Road, Cambridge, CB2 1EZ, UK (fax: +44 1223 336033); on request by e-mail to deposit@ccdc.cam.ac.uk or by access to http://www.ccdc.cam.ac.uk.
References
Dehaen W, Bakulev VA (2014) Topics in heterocyclic Chem, vol 40. Springer, Berlin
Rachwal S, Katritzky AR (2008). In: Katritzy AR, Ramsden CA, Scriven EFV, Taylor RJK eds, Comp Heterocyclic Chem III Ed, 5.01. Pergamon, Oxford, pp 1–158
Bellagamba M, Bencivenni L, Gontrani L, Guidoni L, Sadun C (2013) Struct Chem 4:933–943
Ferreira MLG, Pinheiro LCS, Santos-Filho AO, Peçanha MDS, Sacramento CQ, Machado V, Ferreira VF, Souza TML, Boechat N (2014) Med Chem Res 23:1501–1511
Jordão AK, Afonso PP, Ferreira VF, de Souza MC, Almeida MC, Beltrame CO, Paiva DP, Wardell SMSV, Wardell JL, Tiekink ER, Damaso CR, Cunha AC (2009) Eur J Med Chem 44:37773783
Himanshu H, Tyagi R, Olsen CE, Errington W, Parmar VS, Prasad AK (2002) Biorg Med Chem 10:963–968
Boechat N, Ferreira MLG, Pinheiro LCS, Aguiar AC, Andrade IM, Krettli AU (2014) Chem Biol Drug Des 84:325–332
Ferreira ML, de Souza MVN, Wardell SMSV, Wardell JL, Vasconcelos TRA, Ferreira VF, Lourenço MCS (2010) J Carbohydr Chem 29:265–274
Jordão AK, Sathler PC, Ferreira VF, Campos VR, de Souza MCBV, Castro HC, Lannes A, Lourenco A, Rodrigues CR, Bello ML, Lourenco MCS, Carvalho GSL, Almeida MCB, Cunha AC (2011) Bioorg Med Chem 19:5605–5611
Boechat N, Ferreira VF, Ferreira SB, Ferreira MLG, da Silva FC, Bastos MM, Costa MC, Lourenço MCS, Pinto AC, Krettli AU, Aguiar AC, Teixeira BM, da Silva NV, Martins PRC, Bezerra FAFM, Camilo ALS, da Silva GP, Costa CCP (2011) J Med Chem 54:5988–5999
Lima-Neto RG, Cavalcante NNM, Srivastava RM, Mendonça FJB, Wanderley AG, Neves RP, dos Anjos JV (2012) Molecules 17:5882–5892
da Silva IF, Martins PRC, da Silva EG, Ferreira SB, Ferreira VF, da Costa KRC, de Vasconcellos MC, Lima ES, da Silva FC (2013) Med Chem 9:1085–1090
da Silva FC, de Souza MCBV, Frugulhetti ICPP, Castro HC, Souza SLO, de Souza TML, Rodrigues DQ, Souza AMT, Abreu PA, Passamani F, Rodrigues CR, Ferreira VF (2009) Eur J Med Chem 44:373–383
Weide T, Saldanha SA, Minond D, Spicer TP, Fotsing JR, Spaargaren M, Frere JM, Bebrone C, Sharpless KB, Hodder PS, Fokin VV (2010) ACS Med Chem Lett 1:150–154
Rogawski MA (2006) Epilepsy Res 69:273–294
Jordão AK, Ferreira VF, Souza TML, Faria GGS, Machado V, Abrantes JL, Souza MCBV, Cunha AC (2011) Bioorg Med Chem 19:1860–1965
Shafi S, Alam MM, Mulakayala M, Mulakayala NC, Vanja G, Kalle AM, Pallu R, Alam MS (2012) Eur J Med Chem 49:324–333
Banday AH, Shameem SA, Ganai B (2012) Org Med Chem Lett 2:13
Sumangala V, Poojary B, Chidananda N, Fernandes J, Kumari NS (2010) Arch Pharm ReS 33:1911–1918
Senger MR, Gomes LCA, Ferreira SB, Kaiser CR, Ferreira VF, Paes-Silva F Jr (2012) ChemBioChem 13:1584–1593
Périon R, Ferriéres V, García-Moreno MI, Mellet CO, Duval R, Fernández JMG, Plusquellec D (2005) Tetrahedron 61:9118–9124
Zhou Y, Zhao Y, Boyle KMO, Murphy PV (2008) Bioorg Med Chem Lett 18:954–956
Gonzaga D, Senger MR, da Silva FC, Ferreira VF, Silva FP Jr (2014) Eur J Med Chem 74:461–476
Ferreira VF, da Rocha DR, da Silva FC, Ferreira PG, Boechat NA, Magalhães JL (2013) Expert Opin Ther Pat 23:319–331
Sureshbabu B, Venkatachalam R, Sankararaman S (2014) CrystEngComm 16:6098–6106
Ramana CV, Goriya Y, Durugkar KA, Chatterjee S, Krishnaswamy S, Gonnade RG (2013) CrystEngComm 15:5283–5300
Lumpi L, Glöcklhofer F, Holzer B, Stöger B, Hametner C, Reider GA, Fröhlich J (2014) Cryst Growth Des 14:1018–1031
Kumar AS, Kommu N, Ghuleb VD, Sahoo AK (2014) J Mater Chem A 2:7917–7926
Chevallier F, Blin T, Nagaradja E, Lassagne F, Roisnel T, Halauko YS, Matulis VE, Ivashkevich OA, Mongin F (2012) Org Biomol Chem 10:4878–4885
Liu Y, Yan W, Chen Y, Petersen JL, Shi X (2008) Org Lett 10:5389–5392
Dong ZQ, Liu FM, Zeng YM (2011) J Chem Crystallogr 41:1158–1164
Kamal A, Swapna P (2013) RSC Adv 3:7419–7426
Zhang Y, Wang D, Wang W, Gao T, Wang L, Li J, Huang G, Chen B (2010) Synlett 11:1617–1622
Costa MS, Boechat N, Ferreira VF, Wardell SMSV, Skakle JMS (2006) Acta Crystallogr E62:o1925–o1927
Vinutha N, Kumar SM, Nithinchandra K, Balakrishna K, Lokanath NK, Revannasiddaiah D (2013) Acta Crystallogr E66:o1724
Karimov Z, Abdugafurov I, Talipov S, Tashkhodjaev B (2010) Acta Crystallogr E66:o1674
Costa MS, Boechat N, Rangel EA, da Silva FC, de Souza AMT, Rodrigues CR, Castro HC, Junior IN, Lourenço MCS, Wardell SMSV, Ferreira VF (2006) Biorg Med Chem 14:86448653
Piterskaya YL, Khramchikhin AV, Stadnichuk MD, Bel’sky VK, Stash AL (1996) Zh Obshch Khim 66:1180–1187
Boechat N, Ferreira MLG, Bastos MM, Wardell JL, Wardell SMSV, Tiekink ERT (2011) Acta Crystallogr E67:o2934–o2935
Beyer B, Ulbricht C, Winter A, Hager MD, Hoogenboom R, Herzer N, Baumann SO, Kickelbick G, Görls H, Schubert US (2010) New J Chem 34:2622–2633
Otwinowski Z, Minor W (1997) In: Carter CW Jr, Sweet RM (eds) Methods in enzymology, vol 276, macromolecular crystallography Part A. Academic Press, New York, pp 307–326
Hooft RWW (1998) COLLECT, data collection software. Nonius BV, Delft
Sheldrick GM (2007) SADABS Version 2007/2. Bruker AXS Inc., Madison
Mercury 3.3. Cambridge Crystallographic Data Centre, UK
Sheldrick GM (2008) Acta Crystallogr A 64:112–122
Spek AL (2003) J Appl Crystallogr 36:7–13
Acknowledgments
The use of the NCS crystallographic service at Southampton and the valuable assistance of the staff there are gratefully acknowledged. JLW thanks FAPERJ, Brazil for support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gonzaga, D., da Silva, F.C., Ferreira, V.F. et al. Crystal Structures of 2-Phenyl-2H-1,2,3-Triazol-4-Carbaldehyde, an Active α-Glycosidase Inhibition Agent, and (1-Phenyl-1H-1,2,3-Triazol-4-yl)Methyl Benzoate and (2-(4-Fluorophenyl)-2H-1,2,3-Triazole-4-yl)Methanol, Two Moderately Active Compounds. J Chem Crystallogr 46, 67–76 (2016). https://doi.org/10.1007/s10870-015-0629-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10870-015-0629-4