Abstract
The crystal structures of 4-[(2-hydroxyphenyl)imino]-2-pentanone (H2hpac, 1) and its cobalt(III) complex [CoIII(hpac)py3]+·PF6 − (2) have been determined by X-ray diffraction. The ligand 1 crystallizes in orthorhombic chiral P212121 space group, with a = 8.8405(4) Å, b = 10.5349(8) Å, c = 11.2292)7( Å), and the complex 2—in the centrosymmetric monoclinic P2/n space group, with a = 16.496(5) Å, b = 10.171(2) Å, c = 16.646(5) Å, and β = 95.53(3)°. In the ligand molecule quite strong intramolecular hydrogen bond closes six-membered ring. The bond length pattern within this ring suggests the significant conjugation and the structure might be therefore regarded as the intermediate between keto-enamine and zwitterionic forms, and the intramolecular hydrogen bond falls into category of resonance-assisted hydrogen bonds. In turn, intermolecular O–H···O hydrogen bonds connect the molecules of the ligand into infinite chains along [100] direction. In the complex, the Co(III) ion is hexa-coordinated, by two oxygen and one nitrogen atoms of the doubly-deprotonated ligand 1 and by three nitrogen atoms from three pyridine ligands. The coordination polyhedron is close to a slightly distorted octahedron. The in vitro antimicrobial activity of the Schiff base ligand and its corresponding complex have been tested against human pathogenic bacterias such as Staphylococcus aureus, Bacillus subtilis, Pseudomonas aeruginosa and Escherichia coli.
Graphical Abstract
Resonance-assisted intramolecular hydrogen bond increases the degree of delocalization within the NCCCO fragment of the Schiff base.

Similar content being viewed by others
Results and Discussion
Acetylacetone (acac) or pentane-2,4-dione is often used as a good precursor in organic synthesis and as a building block of metal–organic complexes [1, 2]. Its anion acts as the bidentate ligand and is known to form complexes with many transition metal ions [3–5]. A general method of preparation of such complexes consists in a reaction of a metal ion with acetylacetone in presence of a base which easily separates the proton. Replacement of the ketone functional group by the imine one results in corresponding bidentate Schiff bases, which also proved to be useful ligands. If 2-hydroxy aniline is used as the amine, one can prepare the tridentate ligand. Such a ligand—4-[(2-hydroxyphenyl)imino]-2-pentanone (hereafter referred to as 1), known as H 2 hpac, where H2 represents the dissociable enolic –OH and hydroxyphenyl protons—has been already prepared and used for making the square-planar mixed-ligand nickel(II) complex with deprotonated H 2 hpac (i.e. hpac), which coordinates through the enolate oxygen, the imine nitrogen and the deprotonated hydroxyphenyl oxygen atoms. Additionally, the neutral N-donor, imidazole, has been used as an ancillary ligand [6]. Such compounds are of particular interest due to the asymmetric intramolecular hydrogen bond, formed between oxygen and nitrogen atoms. Depending on the position of the hydrogen atom in this O···H···N bond (which forms a six-membered chelate ring), these o-hydroxy Schiff bases exhibit two tautomeric forms (cf. Scheme 1): the OH form (A) and the NH form (B, C). Additionally, the NH tautomer can exist in the ketoenamine form (B) or the zwitterionic form (C) [7]. The unsaturated –CH=N– bond permits the π-electronic coupling between acidic and basic centres of the molecule. In many o-hydroxy Schiff bases the chelate ring is planar, and is therefore called “pseudoaromatic” chelate ring e.g. [8–10]. An interesting example of such system formed by acac and ethylendiamine fragments, resulting in bis(acetylacetone)ethylenediimine (with an intramolecular ionic hydrogen bond in the solid state) has been reported by Özkar et al. [11].
Cobalt(III) complexes derived from symmetrical and nonsymmetrical Schiff bases have drawn considerable attention in the past for their important biological applications. A number of model complexes with cobalt(II) and cobalt(III) have been prepared with particular emphasis on the reactivity of the metal ions in the transmethylation reaction and on the reversible absorption of molecular oxygen [12–18]. As a continuation of our studies on Schiff bases [19] we have performed the preparation and characterization of [CoIII(hpac)py3]PF6 (hereafter referred to as 2). The spectral properties of the complex were investigated by FT-IR, UV–Vis, and 1HNMR spectra. The X-ray crystal structures of 2 and of the ligand 1 have been determined. The antibacterial activities of both the Schiff base ligand 1 and it’s complex 2 against Bacillus subtilis (Gram-positive), Staphylococcus aureus (Gram-positive), Escherichia coli (Gram-negative) and Pseudomonas aeruginosa (Gram-negative) have also been evaluated.
Synthesis and Characterization
In the preparation of the cobalt(III) complex, a vigorous stream of air was passed through a solution of CoII(hpac) in methanol to which excess pyridine was gradually added. The air oxidation was continued for a period of 3 h during which the color changed from red to green. A solution of KPF6 in methanol was then added to the resulting green solution and stirred for 5 min. Dark red crystals of the complex were obtained in good yield (60 %).
The IR spectra of the free Schiff base ligand and corresponding complex show several bands in the 400–4,000 cm−1 region. The NH stretching frequency of the free ligand, observed at 3,460 cm–1, is due to the internal hydrogen bonding vibration (O−···H–N+). The bands related with the C=O and O–H bonds, present in the free ligand, are not observed in the spectra of the complex. The stretching vibration of PF6 − anion is observed at 841 cm−1 [20]. The electronic absorption spectrum of the complex, measured in chloroform, shows a true maximum band at 575 nm. Relatively low value of ε (181 M−1 cm−1) of this band suggests that it can be assigned to the d–d transition.
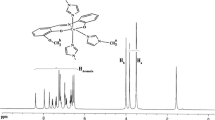
The 1HNMR spectrum of 2 in CDCl3 is shown in Fig. 1 (assignment as in Scheme). These data are in good agreement with the single crystal structure. The expected peaks at 5.32 (s, N–CH2–), 2.58 (s, N=C–CH3), and 2.38 (s, C=C–CH3) ppm are observed. The aromatic protons of the phenyl group appear as a multiplet at the range 6.45–7.19 ppm (Hph), and those of three pyridine molecules—at the range 7.26–8.70 ppm (Hpy).
Molecular Structures
Selected bond lengths and angles for both structures are listed in Table 1.
Figure 2 shows the perspective view of the tridentate Schiff base ligand 1. In the molecule 1 there is a strong intramolecular N–H···O hydrogen bond, with the proton located at the imine nitrogen atom. The position of this hydrogen atom (H8) is proved by finding it in the difference Fourier map and successful refinement, as well as by the geometry of its environment. So far, mainly neutral O–H···N hydrogen bonds have been known in structures of Schiff base ligands [10, 21, 22]. Hydrogen bonding geometry is summarized in Table 2. Analysis of the bond length pattern suggests the significant conjugation within the hydrogen-bonded six-membered ring; in principle the structure might be regarded as the intermediate between B (keto-enamine) and C(zwitterionic) forms in Scheme 1. In a sense then, the structure of H2hpac can be regarded as another example of the ionic hydrogen bonding in a solid Schiff base ligand [7, 9]. The intramolecular ionic hydrogen bonding can be therefore described by employing so-called “resonance assisted hydrogen bonding” (RAHB) model which is essentially a synergetic mutual reinforcement of hydrogen bonding and π-delocalization within the heterodienic system [23]. Here, the delocalization is clearly visible in the bond lengths (cf. Table 1): the C–O bond distance is longer than typical range of double C=O bonds (and compares well with the C–O distance found in the related structure [11]). The C–N bond distance of the azomethine group is longer than a typical C=N bond, but is much shorter than the single C–N bond. Finally, both C–C distances are roughly equal. As another consequence, the six-membered hydrogen-bonded ring is almost planar, with maximum deviation from the least squares plane through all 6 atoms of 0.037(10) Å. The dihedral angle between this ring and the aromatic ring is 30.4(3)°. In the crystal structure the O–H···O intermolecular hydrogen bonds join molecules into infinite chains along [100] direction; these chains are additionally joined by weak C–H···π contacts (Table 2; Fig. 3).
Perspective view of the ligand, H2hpac (1) together with labeling scheme [17]; the displacement ellipsoids are drawn at 50 % probability level, hydrogen atoms are shown as spheres of arbitrary radii. Intramolecular hydrogen bond is drawn as dashed line
The crystal packing of H2hpac as seen along [001] direction [6]. Hydrogen bonds are shown as dashed lines
Figure 4 shows the perspective view of the structure of the complex [CoIII(hpac)py3]PF6. The coordination geometry around the hexa-coordinated Co(III) ion can be described as a slightly distorted octahedron (Table 1). The Co1 ion is coordinated by two oxygen atoms (O42 and O53) and one nitrogen atom (N48) of tridentate Schiff base ligand, and by three nitrogen atoms (N11, N21 and N31) from three pyridine molecules. The trans bond angles of O42–Co1–O53, N11–Co1–N21 and N48–Co1–N31 are 178.38(11)°, 178.66(14)° and 176.59(14)°, respectively, and deviate only slightly from the theoretical value of 180°. All cis bond angles around Co center deviate significantly from 90° indicating a rectangular distortion. Among the cis bond angles, O53–Co1–N48 (95.77(12)°) has the greatest deviation from 90°. The Co—N48 bond is shorter than Co—Npy by approximately 0.08 Å due to the presence of π-backbonding in Co—Nimine bond. The Co–O distances are similar to those found in related complexes e.g. [17, 24]. The geometry of the ligand is in principle similar to that of 1, the dihedral angle between almost planar (within 0.043(2) Å) N=C–C=C–O fragment and the phenyl ring plane is 33.63(12)°. It should be noted however that—in the absence of intramolecular hydrogen bond—the N=C–C=C–O fragment deviates more significantly from the planarity (cf. torsion angles in Table 1). The crystal packing is determined by electrostatic interactions between charged species; some weak C–H··F and C–H···O hydrogen bonds provide additional weak stabilizing forces (Table 2; Fig. 5).
Perspective view of the complex, [CoL(py)3]+PF6 − with labelling scheme [17]. The displacement ellipsoids are drawn at 50 % probability level, hydrogen atoms are shown as spheres of arbitrary radii
The crystal packing of [CoL(py)3]PF6 as seen along [001] direction [33]. The dashed lines denote short contacts (cf. text)
Biological Properties
Potent antibacterial activity of cobalt(II) and cobalt(III) Schiff base complexes have been reported in different studies [25, 26]. Table 3 shows the results of antibacterial activity studies of ligand and its complex evaluated by Kirby–Bauer disc diffusion method against both Gram-positive and Gram-negative bacteria. Although the chemical compounds showed antibacterial activity in disk diffusion assay, their minimal inhibitory concentration (MIC) were more than 30 mg ml−1. It can be concluded that the ligand and its complex had no antibacterial activity against studied bacterial strains.
Experimental
Reagents and Measurements
All other chemicals were commercial reagent grade and used as received from Aldrich and Merck. Elemental analyses were performed by using a Perkin–Elmer 2400II CHNS–O elemental analyzer. UV–Vis spectra were recorded on a JASCO V-570 spectrophotometer. Infrared spectra (KBr pellets) were obtained on a FT-IR JASCO 680 plus spectrophotometer. 1HNMR spectra were obtained on a Bruker Avance DRX 500 (500 MHz) spectrometer. Proton chemical shifts are reported in ppm relative to an internal standard of Me4Si.
Synthesis of Schiff Base Ligand 1
The Schiff base ligand, 4-[(2-hydroxyphenyl)imino]-2-pentanone (H2hpac), was prepared as described in the literature [6]. The single crystals suitable for X-ray data collection were obtained by slow evaporation of the ethanol solution (after 2 days). The crystals were filtered off, washed with a small amount of cold methanol and dried under vacuum.
Synthesis of Complex 2
To a stirring solution of Co(CH3COO)2·4H2O (0.125 g, 0.5 mmol) in methanol (25 ml) was added an equimolar of H2hpac (0.095 g, 0.5 mmol). The pink solution turned brown immediately upon the formation of [CoII(hpac)] complex. To this solution was added 4 mmol of pyridine, and air was bubbled through the reaction mixture for about 3 h. 0.5 mmol of KPF6 was then added to the resulting green brown solution and stirred for 5 min. The single crystals suitable for X-ray data collection were obtained by slow evaporation of the methanol solution after 3 days. The crystals were filtered off,washed with a small amount of cold methanol and dried under vacuum. FT-IR (KBr, cm−1): 1,574 (C=N), 841 (s, PF6) UV–Vis: λmax (nm), ε (L mol−1 cm−1) (CH3CN) 196 (79,000), 275 (70,000), 336 (24,600), 586 (181). 1HNMR (500 MHz, CDCl3, δ, ppm): 2.38 (s, 3H, Ha), 2.58 (s, 3H, Hb), 5.32 (s, 1H, Hc), 6.45–7.19 (4H, phenyl ring proton) 7.26–8.70 (15H, pyridine C–H proton). Anal. Calcd. For C26H26N4O2PF6Co: C, 49.54; H, 4.16; N, 8.89; Found: C, 49.05; H, 4.10; N, 8.68 %.
X-ray Crystallography
Diffraction data for ligand (1) were collected at room temperature by the ω-scan technique on an Agilent Technologies SuperNova four-circle diffractometer with Atlas CCD detector [27], equipped with microfocus CuKα radiation source (λ = 1.54178 Å), while for the complex (2) on an Agilent Technologies Xcalibur four-circle diffractometer with Eos CCD detector [27] and graphite-monochromated MoKα radiation (λ = 0.71069 Å). The data were corrected for Lorentz-polarization as well as for absorption effects [27]. Precise unit-cell parameters were determined by a least-squares fit of 2,198 (1) and 6,083 (2) reflections of the highest intensity, chosen from the whole experiment. The calculations were mainly performed within the WinGX program system [28]. The structures were solved with SIR92 [29] and refined with the full-matrix least-squares procedure on F2 by SHELXL97 [30]. The scattering factors incorporated in SHELXL97 were used. The function ∑w(|Fo|2 − |Fc|2)2 was minimized, with w−1 = [σ 2(F o )2 + (A.P)2 + B.P] (P = [Max (F 2o , 0) + 2F 2c ]/3). The final values of A and B are listed in Table 4. All non-hydrogen atoms were refined anisotropically. All hydrogen atoms from the complex (2) and methyl hydrogen atoms from the ligand (1) were placed in idealized positions and refined as ‘riding model’ with isotropic displacement parameters set at 1.2 (1.5 for methyl groups) times Ueq of appropriate carrier atoms. All other hydrogen atoms from 1 were found in the difference Fourier map and isotropically refined. The crystals of the complex (2) have been found to be twinned, appropriate SHELX refinement procedure has been applied (BASF factor refined at 0.249(1)).
CCDC 860226 (1) and 860227 (2) contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from the Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif or e-mail: deposit@ccdc.cam.ac.uk.
Antibacterial Activity
The antimicrobial activity of synthesized compounds was evaluated by disk diffusion assay [31] and twofold dilution method [32]. The Gram negative and Gram positive standard strains, namely Bacillus subtilis (B. subtilis; PTCC No: 1023; ATCC 6633); Staphylococcus aureus (S. aureus; PTCC No: 1431; ATCC 25923), Escherichia coli (E. coli; PTCC No: 1399; ATCC 25922), and Pseudomonas aeruginosa (P. aeruginosa; PTCC No: 1430; ATCC 27853) were purchased from Iranian Research Organization for Science and Technology (IROST). All bacteria were grown on Muller–Hinton Agar plates (37 °C, 24 h) and the zones of inhibition were measured after 24 h. Each organism was tested in duplicate on different days to measure the reproducibility of the test. Ampicillin, chloramphenicol, kanamycin, and penicillin were purchased from PadtanTeb Company (Iran) and used as reference antibacterial agents.
Conclusion
A new cobalt(III) complex with a tridentate Schiff base ligand and pyridine has been synthesized and characterized by spectroscopic measurements. Structures were determined by X-ray diffraction. The cobalt ion is coordinated by one N and two O atoms from a Schiff base ligand and by the N atoms of three pyridine molecules to form a distorted octahedral geometry. The intramolecular ionic hydrogen bonding is observed in the enolic form of the ligand.
References
Genovese S, Epifano F, Marcotullio MC, Pelucchini C, Curini M (2011) Tetrahedron Lett 52:3474
Bellec N, Massue J, Roisnel T, Lorcy D (2007) Inorg Chem Commun 10:1172
Huq F, Skapski AC (1974) J Cryst Mol Struct 4:411
Von Chrzanowski LS, Lutz M, Spek Al (2007) Acta Crystallogr Sec C Cryst Struct Commun 63:377
Laskar IR, Hsu S-F, Chen T-M (2006) Polyhedron 25:1167
Zhang QL, Zhu BX (2008) J Coord Chem 61:2340
Dominiak PM, Grech E, Barr G, Teat S, Mallinson P, Wozniak K (2003) Chem Eur J 9:963
Filarowski A, Koll A (1998) Vib Spectrosc 17:123
Krygowski TM, Wozniak K, Anulewicz R, Pawlak D, Kolodziejski W, Grech E, Szady A (1997) J Phys Chem A101:9399
Bertolasi V, Gilli P, Ferretti V, Gilli G (1991) J Am Chem Soc 113:4917
Özkar S, Ülkü D, Yıldırım LT, Biricikc N, Gümgüm B (2004) J Mol Struct 688:207
Mondal N, Dey DK, Mitra S, Malik KMA (2000) Polyhedron 19:2707
Parashar RK, Sharma RC, Kumar A, Mohan G (1988) Inorg Chim Acta 151:201
Brückner S, Calligaris M, Nardin G, Randaccio L (1968) Inorg Chim Acta 4:386
Chen H, Han D, Yan H, Tang W, Yang Y, Wang H (1993) Polyhedron 12:1097
Costes JP, Cros G, Darbieu MH, Laurent JP (1982) Inorg Chim Acta 60:111
Böttcher A, Takeuchi T, Hardcastle KI, Meade TJ, Gray HB, Wikel DC, Kapon M, Dori Z (1997) Inorg Chem 36:2498
Clearfield A, Gopal R, Kline RJ, Sipski M, Urban LO (1978) J Coord Chem 7:163
Salehi M, Dutkiewicz G, Kubicki M (2010) Acta Cryst E66:o1590
Amirnasr M, Langer V, Rasouli N, Salehi M, Meghdadi S (2005) Can J Chem 83:2073
Gilli P, Bertolasi V, Ferretti V, Gilli G (1994) J Am Chem Soc 116:909
Wozniak K, He H, Klinowski J, Jones W, Dziembowska T, Grech E (1995) J Chem Soc, Faraday Trans 91:77
Gilli G, Bellucci F, Ferretti V, Bertolasi V (1989) J Am Chem Soc 111:1023
Schenk KJ, Meghdadi S, Amirnasr M, Habibi MH, Amiri A, Salehi M, Kashi A (2007) Polyhedron 26:5448
Nejo AA, Kolawole GA, Nejo AO (2010) J Coord Chem 63:4398
Mishra A, Kaushik NK, Verma AK, Gupta R (2008) Eur J Med Chem 43:2189
Agilent Technologies (2009) CRYSALIS PRO, Version 1.171.33.36
Farrugia LJ (1997) J Appl Crystallogr 30:565
Altomare A, Cascarano G, Giacovazzo C, Gualardi A (1993) J Appl Crystallogr 26:343
Sheldrick GM (2008) Acta Cryst A64:112
Bauer AW, Kirby WM, Sheris JC, Turck M (1966) Am J Clin Pathol 45:493
European Committee for Antimicrobial Susceptibility Testing of the European Society of Clinical Microbiology and Infectious Diseases (2000) Clin Microbiol Infect 6:509
Macrae CF, Bruno IJ, Chisholm JA, Edgington PR, McCabe P, Pidcock E, Rodriguez-Monge L, Taylor R, van de Streek J, Wood PA (2008) J Appl Crystallogr 41:466
Acknowledgments
We thank Semnan University for supporting this study.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Salehi, M., Dutkiewicz, G., Rezaei, A. et al. Synthesis, Antibacterial Studies and Crystal Structures of Tridentate Schiff Base Ligand and It’s Cobalt(III) Complex. J Chem Crystallogr 42, 871–878 (2012). https://doi.org/10.1007/s10870-012-0329-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10870-012-0329-2