Abstract
Resonance assignment is a prerequisite for almost any NMR-based study of proteins. It can be very challenging in some cases, however, due to the nature of the protein under investigation. This is the case with intrinsically disordered proteins, for example, whose NMR spectra suffer from low chemical shifts dispersion and generally low resolution. For these systems, sequence specific assignment is highly time-consuming, so the prospect of using automatic strategies for their assignment is very attractive. In this article we present a new version of the automatic assignment program TSAR dedicated to intrinsically disordered proteins. In particular, we demonstrate how the automatic procedure can be improved by incorporating methods for amino acid recognition and information on chemical shifts in selected amino acids. The approach was tested in silico on 16 disordered proteins and experimentally on α-synuclein, with remarkably good results.
Similar content being viewed by others
Introduction
Nuclear magnetic resonance (NMR) is the most powerful method available for studying intrinsically disordered proteins (IDPs) at atomic resolution. It allows us to obtain a variety of information, including structural propensities, dynamics, and interactions with other molecules. But IDPs are rather difficult objects to study with NMR. The high mobility of the polypeptide chain results in exceptionally narrow ranges of chemical shifts. This effect is amplified by the high incidence of sequential repeats; stretches of three or four residues of the same type are often present in IDPs’ sequences. Also, the high abundance of disorder-promoting amino acids and underrepresentation of others (Dunker et al. 2008) contributes to low chemical shifts dispersion. The usually high content of prolines breaks the chains of sequential connectivities obtained via amide proton detected experiments. The combination of the above factors makes the complete sequence-specific resonance assignment of IDPs a challenging task.
High-dimensional (≥4D) experiments (Kazimierczuk et al. 2013; Nowakowski et al. 2015) enable the spreading of cross-peaks over a larger spectral space, and thus better resolution. However, such techniques require the use of non-uniform sampling (NUS) to accelerate data acquisition. Several methods for processing NUS data have been proposed that make it possible to develop experiments of high dimensionality (Mobli and Hoch 2008; Coggins et al. 2010; Orekhov and Jaravine 2011; Freeman and Kupče 2012; Hiller and Wider 2012; Kazimierczuk et al. 2012; Holland and Gladden 2014). These methods have been applied successfully to IDPs. Efforts have also been made to develop carbon direct-detected techniques (Bermel et al. 2009; Felli and Pierattelli 2014), which are invaluable where the fraction of prolines is high or where amide protons undergo fast chemical exchange (Gil et al. 2013). Carbon direct-detected techniques also show superior chemical shift dispersion compared to amide protons-detected experiments (Brutscher et al. 2015). Moreover, these two strategies can be combined: Several 13C-detected approaches for high-dimensionality have been proposed (Nováček et al. 2011, 2012; Bermel et al. 2012b; Nováček et al. 2013; Bermel et al. 2013; Dziekański et al. 2015).
During the resonance assignment process, the amino acid types of at least some of the residues must be recognized in order to map the chains of sequentially-linked residues onto the polypeptide. Given a single uniformly-labeled protein sample (e.g. without selective isotope labeling), three main methods are used for this purpose: (1) using the statistical values of chemical shifts for various nuclei of different amino acids; (2) using topological information; and (3) using amino acid type selective experiments.
Method (1) is widely used as usually it does not require additional experiments. Cβ and Hβ chemical shifts, which are particularly useful for this purpose, are often recorded within a set of assignment experiments. The statistical values are available from the Biological Magnetic Resonance Bank (BMRB) database (Ulrich et al. 2008), where average Cβ and Hβ chemical shifts for each amino acid are calculated based on at least a few thousand chemical shifts. For IDPs, additional statistics are available (Tamiola et al. 2010), which consider not only the residue type but also the residue’s closest neighbors (i − 1 and i + 1) and are therefore more reliable.
In method (2), the detection of some nuclei limits the range of possible amino acids. For example, the presence of Cβ chemical shift excludes glycine, the presence of HN chemical shift excludes proline, and the presence of two different Hβs excludes alanine, isoleucine, threonine, and valine.
Method (3)—amino acid selective experiments—was first proposed by Dötsch and his coworkers (Dötsch et al. 1996a, b, c; Dötsch and Wagner 1996). The approach is based on the triple-resonance CBCA(CO)NH pulse sequence (Grzesiek and Bax 1992), modified to acquire a signal for certain topology-selected amino acid types. The resulting 2D 1H–15N-HSQC-like spectra contain only resonances originating from the desired amino acid residues.
This concept has since been extensively developed, other researchers adding new selection criteria (Feng et al. 1996; Rios et al. 1996; Schubert et al. 1999, 2000, 2001a, b, c, 2005; Barnwal et al. 2008). The result has been many different strategies, such as the multiplicity selective in-phase coherence transfer (MUSIC) approach developed by Schubert and his collaborators. For selection, several types of pulse sequence components can be employed, including multiple quantum filters (for 13CHn or 15NHn), band-selective pulses on 13C and/or 15N (for specific nuclei excitation), delay tuning (for choosing the desired coherence transfer pathway), and setting an appropriate number of coherence transfer steps (for choosing side-chains of the desired length). Instead of selecting specific correlations, editing can be implemented and combined with the idea of Hadamard encoding to speed up data collection (Lescop et al. 2008; Pantoja-Uceda and Santoro 2008; Lescop and Brutscher 2009; Feuerstein et al. 2012; Pantoja-Uceda and Santoro 2012). More recently, amino acid selection has also been incorporated into 13C-detected experiments (Bertini et al. 2006; Pantoja-Uceda and Santoro 2011; Chakraborty et al. 2012; Jaipuria et al. 2012; Bermel et al. 2012a).
In the current study we show how different amino acid recognition methods can be exploited in automatic resonance assignment, and how the completeness and reliability of the assignment can benefit from this type of information. We present an improved version of the TSAR (Tool for SMFT-based Assignment of Resonances) program (Zawadzka-Kazimierczuk et al. 2012) designed for automatic resonance assignment using experiments of high dimensionality (≥4D). Our improved version includes the information provided by 13C-detected amino acid-selective experiments (Bermel et al. 2012a). Additionally, the IDPs’ chemical shifts’ statistics (Tamiola et al. 2010) are incorporated to enable more efficient chain mapping. Finally, we present a small modification of the 4D HCBCACON pulse sequence (Bermel et al. 2012b) in which peaks of residues possessing a single aliphatic Cγ carbon are of the opposite sign with respect to that of all other residues.
The approach has been tested in simulations on 16 disordered proteins and experimentally on α-synuclein protein, using both 1H-detected (Piai et al. 2014) and 13C-detected experiments (Bermel et al. 2012b, 2013) as a source of sequential correlations. To speed up data collection, all spectra were acquired using NUS, making use of recently developed sampling and processing strategies (Kazimierczuk et al. 2009; Kazimierczuk and Orekhov 2011). Data from the high-dimensional experiments was processed using the sparse multidimensional Fourier transform (SMFT) algorithm (Kazimierczuk et al. 2009), whereas data from the 2D amino acid-selective experiments was processed using the compressed sensing (CS) algorithm (Kazimierczuk and Orekhov 2011).
Materials and methods
All the NMR experiments were performed at 16.4 T on a Bruker Avance spectrometer operating at 700.06 MHz 1H, 176.03 MHz 13C and 70.94 MHz 15N frequencies, equipped with a 13C cryogenically cooled probehead optimized for 13C-direct detection. A sample of 1.0 mM uniformly 13C, 15N labeled human α-synuclein in 20 mM phosphate buffer at pH 6.5 was prepared as previously described (Huang et al. 2005). EDTA and NaCl were added to reach the final concentrations of 0.5 and 200 mM respectively, and 10 % D2O was added for the lock. All experiments were performed at 285.5 K.
The specific parameters for each amino acid selective experiment are reported in the original publication (Bermel et al. 2012a). Those relating to the γ-selective-HCBCACON experiment are given in the legend of the figure describing the pulse sequence (see Figure S1, Supplementary Material). For 13C band-selective π/2 and π flip angle pulses, Q5 (or time reversed Q5) and Q3 shapes (Emsley and Bodenhausen 1992) with durations of 300 and 220 μs respectively were used, except for π pulses that should be band-selective on the Cα region (Q3, 860 μs) and for the adiabatic π pulse to invert both C′ and Cα (smoothed Chirp 500 μs, 25 % smoothing, 80 kHz sweep, 11.3 kHz RF field strength (Bohlen and Bodenhausen 1993)). The 13C band selective pulses on Cali, Cα, and C′ were given at the center of each region, and the adiabatic pulse was adjusted to cover the entire 13C region.
Decoupling of 1H was achieved with waltz16 (Shaka et al. 1983) (1.7 kHz) sequences, and decoupling of 15N with garp4 (Shaka et al. 1985) (1.0 kHz) sequences. Each experiment was performed in a pseudo 2D mode, with States method applied in all indirect dimensions to achieve quadrature detection. All experiments employ the IPAP approach to remove splitting in the direct acquisition dimension caused by the homonuclear Cα–C′ couplings (Bermel et al. 2008).
The experimental parameters are given in Table 1. All experiments were performed using on-grid non-uniform sampling. The “Poisson disk” sampling scheme (Kazimierczuk et al. 2008) was chosen to generate the time schedules with RSPack software. All spectra were acquired using Bruker TopSpin 1.3 software. The experimental data was converted with nmrPipe (Delaglio et al. 1995) and then processed using either the Compressed Sensing (Kazimierczuk and Orekhov 2011) IRLS algorithm with an iteratively changed lp norm (p 1 −> 0) with 30 iterations (2D datasets) or the Sparse Multidimensional Fourier Transform (SMFT) (Kazimierczuk et al. 2009) (4D and 5D datasets) implemented in the Reduced program. Finally, the Sparky program (Goddard and Kneller 2002) was used to display the spectra, and TSAR (Zawadzka-Kazimierczuk et al. 2012) was used to assign the resonances. The RSPack, Reduced and TSAR programs are available free of charge for academic users at http://nmr.cent3.uw.edu.pl/software.
Results and discussion
Methods
The TSAR program (Zawadzka-Kazimierczuk et al. 2012) was developed to analyze data from experiments of high dimensionality processed using the sparse multidimensional Fourier transform (SMFT) algorithm (Kazimierczuk et al. 2009). In this method, instead of computing the full multidimensional spectrum, a set of 2D cross-sections only are calculated. This can be done using the peak list of a lower-dimensional basis spectrum that shares some of the dimensions with the high-dimensional spectrum. For each basis peak, a single cross-section can usually be calculated. Depending on the type of experiment, each cross-section displays one or more peaks; if the experiment provides sequential connectivities, some peaks are redundant in the cross-sections originating from adjacent residues. Importantly, if several multidimensional spectra have to be analyzed together, they must all be processed using the same basis peak list. The strategy for resonance assignment using this kind of input relies on a comparison of the positions of peaks, creating chains of cross-sections. Recognition of the amino acid type of some of the residues makes it possible to map the cross-sections chains onto the protein sequence, which completes the assignment. In the past, TSAR employed just two of the three methods for amino acid identification described in the Introduction to this paper, namely (1) BMRB chemical shift statistics for Cβ, Hβ, Cα, Hα nuclei, and (2) topological information.
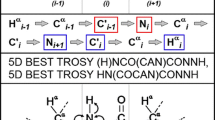
The main goal of the present work was to implement method (3), i.e. amino acid-selective experiments. Previous TSAR version only exploited the change of the sign of peak intensities in the absence of Cβ nucleus for glycine residues, which occurs in experiments where Cα transverse magnetization evolves for c.a. 1/J Cα–Cβ. To see if it was possible to achieve automated assignment of highly overlapping IDP resonances, we decided to use 2D spectra with N and C′ dimensions, which provide superior resolution and make it possible to detect prolines. Two types of such spectra are available (Bermel et al. 2012a): CACON-based and CANCO-based. In the 2D (CA)CON amino acid-selective spectrum, a C′i−1–Ni peak appears if residue i − 1 is of the specified type. In the 2D (CA)NCO amino acid-selective spectrum, a C′i−1−Ni peak appears if residue i − 1 or residue i is of that type. Comparing the two spectra allows us to discriminate C′i−1–Ni peaks related to i − 1 or i residues (Fig. 1).
Analysis of amino-acid selective experiments. As an example, asparagine-selective 2D (CA)CON (left) and (CA)NCO (right) spectra are reported. The basis peak list (black dots) is plotted on top of both spectra. For each C′ i-1 –N i cross-peak, the comparison of the two spectra allows us to determine if asparagine residue is at position i−1 or i
To use such experiments in parallel with SMFT-processed high-dimensional data, the basis peaks corresponding to the selected amino acids must be found. This can be done if the dimensions of the amino acid-selective experiments—in our case amide nitrogen and carbonyl carbon dimensions—are also present in the basis spectrum. The basis peak list can then be plotted on the amino acid-selective spectrum and the basis peaks corresponding to the given amino acid easily identified (Fig. 2). Information on these basis peaks numbers can then be fed into the TSAR program to support the assignment process (for the format of TSAR input files, see Supplementary Material). Of course, it may happen that two basis peaks show up at the position of the amino acid-selective spectrum peak, for example if they are overlapping, or if the amino acid-selective spectrum is not resolved enough. This makes the task more difficult, but TSAR is still able to manage its task. As mentioned above, the only requirement for combining SMFT-processed data with the amino acid-selective data is to have C′ and N dimensions in the basis spectrum. Therefore, although (CA)CON and (CA)NCO experiments exploit carbon detection, they can be combined with both carbon-detected (e.g. with 3D CACON basis spectrum) and also with proton-detected (e.g. with 3D HNCO basis spectrum) experiments for resonance assignment, which makes them even more generally applicable.
Alanine-selective 2D (CA)CON spectrum (red) with the basis peak list superimposed (black dots). On the left, the close-up view of the spectral region inside the blue square is reported. In the example, during the automatic assignment stage, basis peaks on top of the NMR signals are known by TSAR to be related to alanine residues. To make the picture clearer, the number of each basis peak is shown only in the spectral region extracted on the left
In this study we also make use of another method for obtaining amino acid-related information. We modify the 4D HCBCACON pulse sequence (Bermel et al. 2012b) so that the signs of the cross-peaks depend on the topological properties of the residue. The delay for Cβ scalar coupling evolution is increased to 21.0 ms (see Supplementary Material), which allows us to keep the efficiency of the coherence transfer (additional relaxation losses can be neglected for IDPs) and at the same time reverse peak signs for some residues. If a residue i possesses exactly one aliphatic Cγ carbon (E, K, L, M, P, Q, R and T residues), then the H βi –C βi peak has the opposite sign to that of all the other residues. Such zero–one information adds to Cβ and Hβ chemical shifts values, which was the only information provided by the 4D HCBCACON experiment published earlier (Bermel et al. 2012b). It thus improves the performance of the automated assignment. The new pulse sequence also allows us to unambiguously distinguish some residues possessing Cβ and Hβ chemical shifts which may be not so different: S can easily be discriminated from T, I can be easily discriminated from L, and V can easily be discriminated from E, K, M, P, Q, and R. Additionally, the extension of the Cα/β chemical shift evolution increases the resolution of that dimension, with consequent benefits for the comparison of cross-peak spectral positions performed by TSAR. In the old version, TSAR was prepared for single amino acid recognition by peak sign, which was used only for glycine. The current software version accepts sign change in the presence of a user-defined set of residues.
The final major change enabling more efficient amino acid recognition in TSAR is to incorporate the statistical Cα and Cβ chemical shift values calculated using a set of IDPs (Tamiola et al. 2010). TSAR uses statistical chemical shift values at two stages of the operation: (a) recognition of possible amino acids for a single plane, before forming cross-sections chains; and (b) cross-sections chains mapping. During stage (a), the working procedure of TSAR implies that if some chemical shifts exceed the statistical average with four standard deviations for certain amino acids, then these amino acids are excluded from the range of possible ones. During stage (b), if one chain matches in a few sites or a few chains of similar length fit into one site, the deviation from the statistical values in units of standard deviations is calculated, and if the best score is at least three times smaller than the second best, then the better chain is assigned. In the new TSAR version, during stage (b) the chain length is also considered. If up to three Cβ chemical shifts are known, then the better chain is assigned only if the deviation score is at least ten times smaller than the second best.
In the previous software version of TSAR, the BMRB values were used at both stages. In the version of TSAR presented here, the IDP-specific values (Tamiola et al. 2010) are used at stage (b). Such values cannot be employed during stage (a) when the neighboring residues are not yet known, as these values depend on the preceding and following residue type. Thus, at the stage of amino acid recognition for a single plane, the BMRB values are still used. Nonetheless, some IDP-oriented changes were also made at this stage: It was discovered that for IDPs it is better to exclude an amino acid if the chemical shift exceeds two (rather than four) standard deviations. Additionally, Cα chemical shifts were incorporated for amino-acid recognition, while previously only glycine residues could be identified based exclusively on Cα chemical shifts.
Simulations
The new methods of amino acid recognition were tested in a set of simulations. Our aim was to verify the agreement of the statistical values used in the new TSAR version with the real chemical shifts of unstructured proteins. Also, by using the simulations we wanted to check whether the 4D γ-selective—HCBCACON provided information that improves the result of the assignment. An additional goal was to evaluate the effectiveness of incorporating the data from amino-acid selective experiments.
Sixteen proteins (see Table 2) were chosen from the BMRB database. Their lengths ranged from 26 to 467 amino-acid residues. Thirteen of them were natively unstructured, while the remaining three (BMRB IDs 15201, 16626 and 16627) were urea-unfolded proteins. Importantly, none of the proteins chosen by us, except for α-synuclein, was used to prepare the IDPs’ statistics (Tamiola et al. 2010). One of the proteins (BMRB ID 16912) is partially folded, so in the simulations we just used its unstructured C-terminal fragment (residues 79Ser-172Lys). For each of the proteins, peak lists relative to six 13C-detected experiments were generated using the chemical shifts deposited in BMRB. The experiments included 3D CACON, 5D (H)CACON(CA)CON (Bermel et al. 2013), 5D HNCACON (Bermel et al. 2012b), 5D (HCA)CONCACON (Bermel et al. 2013), 4D HCBCACON (Bermel et al. 2012b), and 4D γ-selective-HCBCACON. The peak lists for the 4D and 5D experiments were generated in a format accepted by TSAR: The positions of the peaks in the two dimensions not fixed for SMFT only were given, together with information about the corresponding basis peak. For all proteins, the data from amino-acid selective experiments was also generated.
The artificial data was slightly perturbed: The peak positions were jittered and peak overlap was included. The latter perturbation in particular was realized in three different ways. First, in high-dimensional peak lists the peaks of similar coordinates were joined into a single peak. Second, cross-sections corresponding to overlapping basis peaks were also overlapping: Peaks originating from both overlapping basis peaks appeared on both cross-sections. And third, peak overlap was also considered during the generation of data from amino-acid selective experiments, leading to some ambiguity. The level of perturbations in each of the above aspects was similar to that found for the real IDP data (α-synuclein sample), so the quality of data is similar to the quality of data in the case of real proteins.
In real cases there is an additional source of data imperfection: The dataset is typically incomplete, i.e. some peaks are missing. The level of completeness of the data relates to many factors, including protein concentration, measurement time, pulse sequence efficiency, relaxation rates, and exchange rates. For the proteins used in the simulations, the level of completeness of the data deposited in BMRB ranged from 84.9 to 100 % of the protein residues (excluding the first residue, for which there is no basis peak). The residue was considered to be “present” if all the chemical shifts of the corresponding basis peak were known. As a result, for some “present” residues, certain resonances were unknown.
Several datasets were constructed for each protein. Each dataset included the basis spectrum 3D CACON and one, two or three 5D spectra providing the sequential connectivities. Various numbers of linking experiments were used, due to the fact that the TSAR program forms cross-sections chains whose lengths depend on the quality of the experiments providing sequential correlations. Thus, the datasets containing different combinations of such experiments allow us to evaluate the efficiency of amino-acid recognition methods for various chain lengths. In datasets A1–A6, the linking experiment was 5D (H)CACON(CA)CON. In datasets B1–B6, they were 5D (H)CACON(CA)CON and 5D HNCACON. In datasets C1–C6, the connectivities were provided by 5D (H)CACON(CA)CON, 5D HNCACON, and 5D (HCA)CONCACON spectra. For protein 19135, due to the lack of information about HN chemical shifts in the BMRB entry, the experiments providing the sequential connectivities were (H)CACON(CA)CON and (HCA)CONCACON for datasets B1–B6. Datasets C1–C6 were not constructed in this case.
For amino acid recognition, in some datasets either the 4D HCBCACON (Bermel et al. 2012b) or the new 4D γ-selective-HCBCACON experiments yielding Cβ and Hβ chemical shifts was employed, while in others the information provided by the amino acid-selective experiments was used. The latter group consisted of 13C-detected 2D (CA)CON- and (CA)NCO-based amino acid selective experiments (Bermel et al. 2012a), including the following selections: A, D, E, FHYW, G, N, Q, and S. Datasets A1, B1, and C1 did not contain any additional information on amino acids. Datasets A2, B2 and C2 exploited the 4D HCBCACON experiment. Datasets A3, B3, and C3 used the 4D γ-selective-HCBCACON experiment, which carries extra information in the peak signs. Datasets A4, B4, and C4 employed all eight amino-acid selective 2D experiments (selecting A, D, E, G, N, Q, S, and FHYW), both in the (CA)CON and (CA)NCO versions. Datasets A5, B5, and C5 employed all 2D amino acid selective experiments, but in the (CA)CON version only. Datasets A6, B6, and C6 exploited some of the amino-acid selective experiments in both the (CA)CON and (CA)NCO versions. The choice of amino-acids to be selected was based on the sequence of each protein (see Table 2), such that approximately 40 % of residues were extracted from the total. This meant that the numbers of aa-selective experiments varied from two (as for protein 16912) to five (as for protein 15883), depending on the abundances of different amino acids in a given protein.
The datasets were analyzed using the new version of the TSAR program, and the results compared with the original BMRB assignment. Additionally, datasets A1, B1, C1, A2, B2, and C2 were analyzed using the old version of the TSAR program, to compare the efficiency of the amino-acid recognition procedures in the old and new versions. Datasets 3, 4, 5, and 6 are not accepted by the old version of TSAR. The results are presented in Table 3 (datasets A1–A6), Table 4 (datasets B1–B6), and Table 5 (datasets C1–C6).
To assess the data, we need to consider the lengths of the cross-sections chains formed by TSAR. The longer the chain, the easier and more reliable its mapping onto the protein sequence. Chains were divided into two groups: long (≥4 cross-sections) and short (1–3 cross-sections). The length of the chains is influenced by several factors—not just the protein size, but also the chemical shift dispersion, the number of missing basis peaks, the number and quality of connectivity-yielding experiments, and the number of prolines (in the case of HN-detected experiments). As can be seen by comparing datasets A, B, and C, the proportion of cross-sections within the long chains generally increases in line with the number of linking experiments (see Tables 3, 4 and 5). This reflects different levels of assignment difficulty for the datasets A, B, and C.
Another factor also influences the complexity of the assignment process: the incidence of repeats in the sequence of proteins. This includes overrepresentation of certain amino acid in the sequence, stretches of several residues of the same amino acids, and multiple occurrences of certain sequential motifs. This factor is more difficult to measure, but should be considered when interpreting the data. In this respect, protein 18851, which contains 11 Arg residues within its 26-residues-long sequence (including one four-Arg stretch), is considered difficult to assign despite its small size.
Several conclusions can be drawn from the resulting data. First, it is evident that providing any type of information on amino acids significantly improves the result of the assignment (datasets 2, 3, 4, 5, and 6 vs. datasets 1). This is not surprising, as the mapping of the cross-sections chains onto the protein sequence is performed using amino acid recognition. For instance, for the protein 15179, the result for dataset A1 is 29.7 % correct assignments, which can be increased to 84.2 % correct assignments for the dataset A4. Where there are only a few cross-sections in short chains (typically below 10 %), they can often be correctly mapped onto the sequence even without the additional information on amino acids. This is true for protein 15883, for example, where 95.6 % correct assignments were obtained for all datasets B1–B6, the maximum possible taking into consideration the completeness of the data. If the chains are very short (typically over 40 % of cross-sections within short chains), extra information on amino acids is beneficial, but the results may still not be satisfactory. This is true for protein 16912 datasets A, for example, where the result can be improved from 7.5 % (A1) to 35.5 % (A4), but is still much too low. In such cases, more experiments providing the sequential connectivities are required (see datasets B and C for protein 16912).
Comparing the results obtained with the old and new versions of TSAR (datasets A1, B1, C1, A2, B2, and C2), we find that the new version generates more reliable results: The proportion of incorrect assignments is significantly lower. This improvement is due to the more careful assignment of short chains. In many cases, introducing stricter rules reduces the numbers of both correct and incorrect assignments of short chains (see, for example, protein 16445 datasets A2 and B2). This is both safe and beneficial: Even if some short chains remain unassigned, reducing the number of errors is advantageous. At the same time, long chains are assigned more efficiently by employing the chemical shift statistics for IDPs (see, for example, protein 19135 dataset B2 and protein 15176 dataset C2). The positive effect of incorporating Cα chemical shift statistics for non-glycine residues is reflected in the significant improvement in program’s performance for datasets (A1, B1, C1).
The datasets A for protein 15201 allow us to verify the usefulness of the approach—in particular the IDPs statistics—for urea-unfolded proteins. A total of 87.8 % of residues were correctly assigned for datasets A2 and A3, compared to just 33.3 % for dataset A1. This indicates that the statistical values correctly reflect the chemical shifts of this urea-unfolded protein. In two other urea-unfolded proteins (16626 and 16627), the cross-sections chains were so long that even for dataset A1 almost complete assignment was obtained. For protein 16627, the aliphatic chemical shifts appear to be consistent with the statistics used by TSAR, but for protein 16626 we observed a reduction in the proportion of correct assignments by 1.6 percentage points for datasets containing aliphatic chemical shifts (A2, A3, B2, B3, C2, C3) compared to those lacking such information. The reason for this was the exclusion of the correct amino acid (Asp) from the set of possibilities for one residue. This happened during the first step of amino acid recognition: The Cβ chemical shift slightly exceeded the range for Asp (BMRB average ± 2 SD). Importantly, the mismatch between the statistics and the real data does not result in incorrect assignment. Overall, therefore, the procedures proposed in this article appear to be applicable for urea-unfolded proteins.
The next question examined during the simulations concerned the amount of information from the 4D γ-selective-HCBCACON experiment versus the 4D HCBCACON. In most cases, the result from datasets A2, B2, and C2 was identical to the results from A3, B3, and C3 respectively. However, in several cases the result was better when using the new γ-selective experiment, inasmuch as the number of correct assignments rose or the number of incorrect assignments fell (e.g. 11526 or 18895 datasets A2–A3). In only one case did replacing the HCBCACON with the γ-selective-HCBCACON decrease the number of correct assignments, namely protein 18580 datasets A2–A3. At the same time, it also removed the incorrect assignment. Overall, therefore, we recommend using the γ-selective-HCBCACON rather than the HCBCACON as it yields extra information without taking extra time.
The simulations indicate that it is usually beneficial to employ amino-acid selective experiments in place of—or in addition to—aliphatic chemical shifts. In twenty-four cases, the result was better for dataset 4 than for the corresponding dataset 3; in only six cases did the opposite apply. In two cases, however, using amino-acid selective experiments as the only source of information on amino acids caused problems. The first case was that of protein 18417 datasets B4 and B5, where an incorrect chain of two cross-sections was mapped. The second case was that of protein 19135 dataset A4, where an incorrect chain constructed of five cross-sections (with a single incorrect link inside it) was mapped. However, in this second case over 68 % of cross-sections were within short chains and the overall result was very low (around 60 % of assigned residues), thus it was a ‘high-risk’ dataset. Overall, therefore, using amino acid-selective experiments appears to be a reliable alternative. Of course, if Cβ and Hβ chemical shifts are essential for further studies, the method of choice would be the 4D HCBCACON experiment. But if only the backbone assignment is desired, it is worth considering collecting amino acid-selective 2D spectra.
As shown by datasets 5 and 6, the completeness of the resonance assignment falls when the amount of amino acid-selective data is decreased, although the longer the cross-sections chains, the less visible this effect. The proportion of cross-sections in short chains is therefore a very important parameter for suggesting how many amino acid selective experiments are worth acquiring. Clearly, when making the selections for a particular sample, the protein’s sequence should be considered: It is more valuable to identify the amino acids that are abundant in a given molecule. By comparing datasets 5 and 6, we hoped to identify whether it is more beneficial to acquire more amino-acid selective experiments but only in the CACON version (datasets 5), or fewer selections but in both the CACON and the CANCO versions (datasets 6). Regrettably, no definite conclusion could be drawn here: In seven cases the result was higher for dataset 5, and in eight cases it was higher for dataset 6.
Overall, the results are satisfactory. For 11 proteins, over 95 % of residues whose chemical shifts were deposited in BMRB were correctly assigned. Only in the case of one protein was the proportion below 90 %. Incorrect assignments were rare: They occurred for seven proteins in datasets A (with one linking experiment), four proteins in datasets B (with two linking experiments), and two proteins in datasets C (with three linking experiments). Importantly, the assignment result can usually be increased manually: TSAR provides information that allows the user to get back to doubtful fragments of spectra easily and complete the assignment process. At the same time, incorrect assignments can be identified; a manual inspection is always recommended for short chains. In the simulations, almost all incorrect assignments occurred for short chains or at the very end of long chains, which is relatively easy to spot during manual inspection of the result. The only cases of incorrect assignment for long chains were protein 19135 dataset A4, as mentioned above, and protein 11526 dataset A2 and A5 (4 cross-sections-long chain). In the case of protein 11526, the reason was the very untypical (56.52 ppm) Cα chemical shift of 21Val residue.
Experimental results
The new methods of amino acid recognition were also tested on α-synuclein protein, using 13C- or 1H-detected basis and high-dimensional experiments. In the case of 13C-detected data, amino acid recognition was achieved by using 4D HCBCACON or 4D γ-selective-HCBCACON experiments, or by using the 2D amino-acid selective experiments (eight selections: A, D, E, G, N, Q, S, and FHYW, each in both the (CA)CON and (CA)NCO versions). For 1H-detected data, amino acid recognition was achieved only using 2D amino-acid selective experiments; no experiment providing Cβ and Hβ chemical shifts was acquired in this case.
The selectivity of the amino acid-selective experiments is reported in Table S1 in the Supplementary Material. In all the spectra, only the peaks of the selected amino acid (or amino acids) are present, with few exceptions: In Q-selective 2D (CA)CON and 2D (CA)NCO experiments there is a leakage of N peaks, but in the 2D (CA)NCO experiment they have the opposite sign to the Q peaks and so are very easy to recognize; in E-selective 2D (CA)CON experiment, D peaks appear, but with opposite sign to the E peaks; in G- and S-selective and 2D (CA)NCO experiments, peaks originating form P residues are present, but in the G-selective one they have the opposite sign to the other peaks. TSAR was thus trained to handle such cases. Regarding the completeness of the information, all the expected peaks were retrieved for all the amino acid-selective spectra.
Twelve datasets were constructed out of the 13C-detected data (Table 6). Datasets A1–A4 contain only one links-yielding 5D (H)CACON(CA)CON experiment. Datasets B1–B4 include the 5D (H)CACON(CA)CON and 5D HNCACON experiments. Datasets C1–C4 comprise the 5D (H)CACON(CA)CON, 5D HNCACON, and 5D (HCA)CONCACON experiments. Datasets A1, B1, and C1 do not contain any additional information on amino acids. Datasets A2, B2, and C2 use the 4D HCBCACON experiment. Datasets A3, B3, and C3 use the 4D γ-selective-HCBCACON experiment. Datasets A4, B4, and C4 use all 2D amino acid selective experiments, both in the (CA)CON and (CA)NCO versions.
The results obtained by the TSAR program for different datasets of 13C-detected experiments are presented in Table 7. The data analysis shows the effectiveness of the approach. Even using a single experiment yielding the sequential connectivities, 83.5 % correct assignments were achieved when combined with amino acid-selective data. Using two linking experiments made it possible to achieve 86.3 % correct assignments and 2.2 % incorrect ones, using a very limited amount of information on amino acid types (glycine recognition based on the peaks’ signs and the CA chemical shifts). Incorporating additional information (CB chemical shifts or amino-acid selective experiments) improved this result further still. For datasets C1–C4, the result was 89.9 % correct assignments and no incorrect ones.
Datasets A1, B1, C1, A2, B2, and C2 allowed us to compare the performance of the old and the new versions of the TSAR program (data from the other datasets could not be processed by the old version). As with the simulation, the results demonstrate that the new version of TSAR performs better than the old one: In particular, the number of incorrect assignments was lower in four of the datasets. Only in one dataset (B1) the new program yield more incorrect assignments than the old one (2.2 vs. 0.7 %), but here the number of correct assignments was significantly higher (86.3 vs. 70.5 %).
Using a γ-selective-HCBCACON experiment (datasets A3, B3, C3) instead of the standard HCBCACON experiment (datasets A2, B2, C2) does not cause a significant change in the assignment results. For datasets exploiting one linking experiment only, it allows us to significantly increase the number of correct assignments, but at the same time increases the number of errors (datasets A3 vs. A2).
Using amino acid-selective experiments (datasets A4, B4, C4, D4) seems to be a reliable alternative to measuring Cβ and Hβ chemical shifts for amino acid recognition. In the case of shorter cross-sections chains, TSAR performs even better than using β chemical shifts (dataset A4 vs. A2 and A3). Nonetheless, for dataset B4 it introduces some erroneous assignments (in short chains and one at the end of a long chain). It is striking that the total experimental time required for a set of 2D amino acid-selective experiments less than half that required for the HCBCACON (12 hours, compared to 28 hours; see Table 6).
Alternatively, 1H-detected experiments providing sequential connectivities can be used for assignment, e.g. 5D BT-HN(COCAN)CONNH and 5D BT-(H)NCO(CAN)CONNH experiments (Piai et al. 2014), which require a 3D BT-HNCO as basis spectrum. Eighteen datasets featuring such experiments were constructed in our study (Table 8). Datasets D1–D6 contain the 5D BT-HN(COCAN)CONNH experiment; datasets E1–E6 include the 5D BT-(H)NCO(CAN)CONNH experiment; and datasets F1–F6 contain both experiments. Different datasets contain different combinations of amino-acid selection experiments. Thus, datasets D1, E1, and F1 include all selections (A, D, E, FHYW, G, N, Q, S) in both the (CA)CON and (CA)NCO versions; datasets D2, E2, and F2 contain the same selections but only in the (CA)CON version; datasets D3, E3, and F3 comprise A, E, G, FHYW, and Q selections in both the (CA)CON and (CA)NCO versions; datasets D4, E4, and F4 include A, E, G, FHYW, and Q selections but only in the (CA)CON version; datasets D5, E5, and F5 include A, E, and G selections in both the (CA)CON and (CA)NCO versions; and datasets D6, E6, and F6 contain A, E, and G selections, but only in the (CA)CON version.
The results provided by the TSAR program using these datasets are summarized in Table 9. Here, again, the robustness of the assignment procedure is confirmed. While using all amino acid selection experiments, the percentage of correct assignments obtained using just a single 5D experiment to establish sequential correlations was 79.9 % for BT-HN(COCAN)CONNH (dataset D1) and 88.8 % for BT-(H)NCO(CAN)CONNH (dataset E1). It is worth mentioning that incorrect assignment (one residue within a short chain) occurred only for datasets A, where the cross-sections chains were shortest.
Conclusions
In this study we present a refined version of the automatic resonance assignment TSAR program, with improved assignment efficiency and reliability for IDPs. The changes were made with particular objectives in mind: to exploit peaks’ signs depending on the originating residue; to employ the chemical shift statistics established especially for IDPs; and to analyze the data from amino acid-selective experiments. Besides improving the TSAR program, we also propose a modification of an existing 4D HCBCACON experiment so that information on the amino acid type is coded in the peak sign. These methods were tested in simulations using 16 disordered proteins from the BMRB data base, and then verified experimentally using α-synuclein, a 140-amino acids-long IDP, for both proton- and carbon-detected experiments.
The analysis shows that incorporating the above methods significantly improves the results of the assignment, especially for datasets in which the cross-sections chains are relatively short. Amino acid-selective experiments, which are relatively quick when performed using non-uniform sampling, can be used as an alternative to amino acid recognition based on chemical shift analysis. The proposed methods facilitate the resonance assignment of IDPs and make it both more reliable and more complete.
References
Barnwal RP, Rout AK, Atreya HS, Chary KVR (2008) Identification of C-terminal neighbours of amino acid residues without an aliphatic 13Cγ as an aid to NMR assignments in proteins. J Biomol NMR 41:191–197. doi:10.1007/s10858-008-9254-2
Bermel W, Felli IC, Kummerle R, Pierattelli R (2008) 13C direct-detection biomolecular NMR. Concepts Magn Reson A 32A:183–200. doi:10.1002/cmr.a
Bermel W, Bertini I, Felli IC, Pierattelli R (2009) Speeding up 13C direct detection biomolecular NMR spectroscopy. J Am Chem Soc 131:15339–15345. doi:10.1021/ja9058525
Bermel W, Bertini I, Chill J, Felli IC, Haba N, Kumar MVV, Pierattelli R (2012a) Exclusively heteronuclear 13C-detected amino-acid-selective NMR experiments for the study of intrinsically disordered proteins (IDPs). ChemBioChem 13:2425–2432. doi:10.1002/cbic.201200447
Bermel W, Bertini I, Felli IC, Gonnelli L, Koźmiński W, Piai A, Pierattelli R, Stanek J (2012b) Speeding up sequence specific assignment of IDPs. J Biomol NMR 53:293–301. doi:10.1007/s10858-012-9639-0
Bermel W, Felli IC, Gonnelli L, Koźmiński W, Piai A, Pierattelli R, Zawadzka-Kazimierczuk A (2013) High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J Biomol NMR 57:353–361. doi:10.1007/s10858-013-9793-z
Bertini I, Felli IC, Gonnelli L, Pierattelli R, Spyranti Z, Spyroulias GA (2006) Mapping protein-protein interaction by 13C-detected heteronuclear NMR spectroscopy. J Biomol NMR 36:111–122. doi:10.1007/s10858-006-9068-z
Bohlen JM, Bodenhausen G (1993) Experimental aspects of chirp NMR Spectroscopy. J Magn Reson Ser A 102:293–301. doi:10.1006/jmra.1993.1107
Brutscher B, Felli IC, Gil-Caballero S, Hošek T, Kümmerle R, Piai A, Pierattelli R, Sólyom Z (2015) NMR Methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: general overview and practical guidelines. Adv Exp Med Biol 870:49–122. doi:10.1007/978-3-319-20164-1_3
Chakraborty S, Susmitha AL, Hosur RV (2012) Selective lighting up of segments around Gly, Ala and Ser/Thr in proteins. Magn Reson Chem 50:587–591. doi:10.1002/mrc.3843
Coggins BE, Venters RA, Zhou P (2010) Radial sampling for fast NMR: concepts and practices over three decades. Prog Nucl Magn Reson Spectrosc 57:381–419. doi:10.1016/j.pnmrs.2010.07.001
Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. doi:10.1007/BF00197809
Dötsch V, Wagner G (1996) Editing for amino-acid type in CBCACONH experiments based on the 13C beta-13C gamma coupling. J Magn Reson B 111:310–313. doi:10.1006/jmrb.1996.0100
Dötsch V, Matsuo H, Wagner G (1996a) Amino-acid-type identification for deuterated proteins with a beta-carbon-edited HNCOCACB experiment. J Magn Reson B 112:95–100. doi:10.1006/jmrb.1996.0117
Dötsch V, Oswald RE, Wagner G (1996b) Selective identification of threonine, valine, and isoleucine sequential connectivities with a TVI-CBCACONH experiment. J Magn Reson B 110:304–308. doi:10.1006/jmrb.1996.0047
Dötsch V, Oswald RE, Wagner G (1996c) Amino-acid-type-selective triple-resonance experiments. J Magn Reson B 110:107–111
Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN (2008) The unfoldomics decade: an update on intrinsically disordered proteins. BMC Gen 9(Suppl 2):S1. doi:10.1186/1471-2164-9-S2-S1
Dziekański P, Grudziąż K, Jarvoll P, Koźmiński W, Zawadzka-Kazimierczuk A (2015) 13C-detected NMR experiments for automatic resonance assignment of IDPs and multiple-fixing SMFT processing. J Biomol NMR 62:179–190. doi:10.1007/s10858-015-9932-9
Emsley L, Bodenhausen G (1992) Optimization of shaped selective pulses for NMR using a quaternion description of their overall propagators. J Magn Reson 97:135–148. doi:10.1016/0022-2364(92)90242-Y
Felli IC, Pierattelli R (2014) Novel methods based on 13C detection to study intrinsically disordered proteins. J Magn Reson 241:115–125. doi:10.1016/j.jmr.2013.10.020
Feng W, Rios CB, Montelione GT (1996) Phase labeling of C-H and C-C spin-system topologies: application in PFG-HACANH and PFG-HACA(CO)NH triple-resonance experiments for determining backbone resonance assignments in proteins. J Biomol NMR 8:98–104
Feuerstein S, Plevin MJ, Willbold D, Brutscher B (2012) IHADAMAC: a complementary tool for sequential resonance assignment of globular and highly disordered proteins. J Magn Reson 214:329–334. doi:10.1016/j.jmr.2011.10.019
Freeman R, Kupče E (2012) Concepts in projection-reconstruction. Top Curr Chem 316:1–20. doi:10.1007/128
Gil S, Hošek T, Solyom Z, Kümmerle R, Brutscher B, Pierattelli R, Felli IC (2013) NMR spectroscopic studies of intrinsically disordered proteins at near-physiological conditions. Angew Chem Int Ed Engl 52:11808–11812. doi:10.1002/anie.201304272
Goddard TD, Kneller DG (2002) Sparky 3. University of California, San Francisco
Grzesiek S, Bax A (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc 114:6291–6293. doi:10.1021/ja00042a003
Hiller S, Wider G (2012) Automated projection spectroscopy and its applications. Top Curr Chem 316:21–47. doi:10.1007/128
Holland DJ, Gladden LF (2014) Less is more: how compressed sensing is transforming metrology in chemistry. Angew Chem Int Ed 53:13330–13340. doi:10.1002/anie.201400535
Huang C, Ren G, Zhou H, Wang C (2005) A new method for purification of recombinant human alpha-synuclein in Escherichia coli. Protein Expr Purif 42:173–177. doi:10.1016/j.pep.2005.02.014
Jaipuria G, Lobo NP, Shet D, Atreya HS (2012) High resolution methyl selective 13C-NMR of proteins in solution and solid state. J Biomol NMR 54:33–42. doi:10.1007/s10858-012-9647-0
Kazimierczuk K, Orekhov VY (2011) Accelerated NMR spectroscopy by using compressed sensing. Angew Chem Int Ed Engl 50:5556–5559. doi:10.1002/anie.201100370
Kazimierczuk K, Zawadzka A, Koźmiński W (2008) Optimization of random time domain sampling in multidimensional NMR. J Magn Reson 192:123–130. doi:10.1016/j.jmr.2008.02.003
Kazimierczuk K, Zawadzka A, Koźmiński W (2009) Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J Magn Reson 197:219–228. doi:10.1016/j.jmr.2009.01.003
Kazimierczuk K, Misiak M, Stanek J, Zawadzka-Kazimierczuk A, Koźmiński W (2012) Generalized Fourier Transform for non-uniform sampled data. Top Curr Chem 316:79–124. doi:10.1007/128
Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Koźmiński W (2013) High-dimensional NMR spectra for structural studies of biomolecules. ChemPhysChem 14:3015–3025. doi:10.1002/cphc.201300277
Lescop E, Brutscher B (2009) Highly automated protein backbone resonance assignment within a few hours: The “BATCH” strategy and software package. J Biomol NMR 44:43–57. doi:10.1007/s10858-009-9314-2
Lescop E, Rasia R, Brutscher B (2008) Hadamard amino-acid-type edited NMR experiment for fast protein resonance assignment. J Am Chem Soc 130:5014–5015. doi:10.1021/ja800914h
Mobli M, Hoch JC (2008) Maximum entropy spectral reconstruction of non-uniformly sampled data. Concepts Magn Reson A Bridg Educ Res 32A:436–448. doi:10.1002/cmr.a.20126
Nováček J, Zawadzka-Kazimierczuk A, Papoušková V, Žídek L, Sanderová H, Krásný L, Koźmiński W, Sklenář V (2011) 5D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion. J Biomol NMR 50:1–11. doi:10.1007/s10858-011-9496-2
Nováček J, Haba NY, Chill JH, Zídek L, Sklenář V (2012) 4D non-uniformly sampled HCBCACON and 1J(NCα)-selective HCBCANCO experiments for the sequential assignment and chemical shift analysis of intrinsically disordered proteins. J Biomol NMR 53:139–148. doi:10.1007/s10858-012-9631-8
Nováček J, Janda L, Dopitová R, Žídek L, Sklenář V (2013) Efficient protocol for backbone and side-chain assignments of large, intrinsically disordered proteins: transient secondary structure analysis of 49.2 kDa microtubule associated protein 2c. J Biomol NMR 56:291–301. doi:10.1007/s10858-013-9761-7
Nowakowski M, Saxena S, Stanek J, Żerko S, Koźmiński W (2015) Applications of high dimensionality experiments to biomolecular NMR. Prog Nucl Magn Reson Spectrosc 90–91:49–73. doi:10.1016/j.pnmrs.2015.07.001
Orekhov VY, Jaravine VA (2011) Analysis of non-uniformly sampled spectra with multi-dimensional decomposition. Prog Nucl Magn Reson Spectrosc 59:271–292. doi:10.1016/j.pnmrs.2011.02.002
Pantoja-Uceda D, Santoro J (2008) Amino acid type identification in NMR spectra of proteins via β- and γ-carbon edited experiments. J Magn Reson 195:187–195. doi:10.1016/j.jmr.2008.09.010
Pantoja-Uceda D, Santoro J (2011) Selective observation of Asp and Glu resonances in 13CO detected experiments. Magn Reson Chem 49:558–561. doi:10.1002/mrc.2780
Pantoja-Uceda D, Santoro J (2012) New amino acid residue type identification experiments valid for protonated and deuterated proteins. J Biomol NMR 54:145–153. doi:10.1007/s10858-012-9665-y
Piai A, Hošek T, Gonnelli L, Zawadzka-Kazimierczuk A, Koźmiński W, Brutscher B, Bermel W, Pierattelli R, Felli IC (2014) “CON-CON” assignment strategy for highly flexible intrinsically disordered proteins. J Biomol NMR 60:209–218. doi:10.1007/s10858-014-9867-6
Rios CB, Feng W, Tashiro M, Shang Z, Montelione GT (1996) Phase labeling of C-H and C-C spin-system topologies: application in constant-time PFG-CBCA(CO)NH experiments for discriminating amino acid spin-system types. J Biomol NMR 8:345–350. doi:10.1007/BF00410332
Schubert M, Smalla M, Schmieder P, Oschkinat H (1999) MUSIC in triple-resonance experiments: amino acid type-selective (1)H–(15)N correlations. J Magn Reson 141:34–43. doi:10.1006/jmre.1999.1881
Schubert M, Ball LJ, Oschkinat H, Schmieder P (2000) Bridging the gap: a set of selective 1H-15N-correlations to link sequential neighbors of prolines. J Biomol NMR 17:331–335. doi:10.1023/A:1008362904205
Schubert M, Oschkinat H, Schmieder P (2001a) MUSIC, selective pulses, and tuned delays: amino acid type-selective 1H–15N correlations, II. J Magn Reson 148:61–72. doi:10.1006/jmre.2000.2222
Schubert M, Oschkinat H, Schmieder P (2001b) MUSIC and aromatic residues: amino acid type-selective 1H–15N correlations, III. J Magn Reson 153:186–192. doi:10.1006/jmre.2001.2447
Schubert M, Oschkinat H, Schmieder P (2001c) Amino acid type-selective backbone 1H–15N-correlations for Arg and Lys. J Biomol NMR 20:379–384
Schubert M, Labudde D, Leitner D, Oschkinat H, Schmieder P (2005) A modified strategy for sequence specific assignment of protein NMR spectra based on amino acid type selective experiments. J Biomol NMR 31:115–128. doi:10.1007/s10858-004-8263-z
Shaka A, Keeler J, Freeman R (1983) Evaluation of a new broadband decoupling sequence: WALTZ-16. J Magn Reson 53:313–340. doi:10.1016/0022-2364(83)90035-5
Shaka A, Barker P, Freeman R (1985) Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson 64:547–552. doi:10.1016/0022-2364(85)90122-2
Tamiola K, Acar B, Mulder FAA (2010) Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc 132:18000–18003. doi:10.1021/ja105656t
Ulrich EL, Akutsu H, Doreleijers JF et al (2008) BioMagResBank. Nucleic Acids Res 36:D402–D408. doi:10.1093/nar/gkm957
Zawadzka-Kazimierczuk A, Koźmiński W, Billeter M (2012) TSAR: a program for automatic resonance assignment using 2D cross-sections of high dimensionality, high-resolution spectra. J Biomol NMR 54:81–95. doi:10.1007/s10858-012-9652-3
Acknowledgments
This study was supported by Grant No. IP2012 062772 from the Polish Ministry of Science and Higher Education for the years 2013–2016, by the POMOST program of the Foundation for Polish Science, and by the European Commission Projects IDPbyNMR (Contract No. 264257), BioNMR (Contract No. 261863) and INSTRUCT (Contract No. 211252).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Piai, A., Gonnelli, L., Felli, I.C. et al. Amino acid recognition for automatic resonance assignment of intrinsically disordered proteins. J Biomol NMR 64, 239–253 (2016). https://doi.org/10.1007/s10858-016-0024-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-016-0024-2