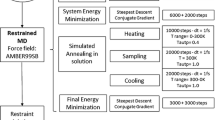
Abstract
Correctly calculating the structure of metal coordination sites in a protein during the process of nuclear magnetic resonance (NMR) structure determination and refinement continues to be a challenging task. In this study, we present an accurate and convenient means by which to include metal ions in the NMR structure determination process using molecular dynamics (MD) simulations constrained by NMR-derived data to obtain a realistic and physically viable description of the metal binding site(s). This method provides the framework to accurately portray the metal ions and its binding residues in a pseudo-bond or dummy-cation like approach, and is validated by quantum mechanical/molecular mechanical (QM/MM) MD calculations constrained by NMR-derived data. To illustrate this approach, we refine the zinc coordination complex structure of the zinc sensing transcriptional repressor protein Staphylococcus aureus CzrA, generating over 130 ns of MD and QM/MM MD NMR-data compliant sampling. In addition to refining the first coordination shell structure of the Zn(II) ion, this protocol benefits from being performed in a periodically replicated solvation environment including long-range electrostatics. We determine that unrestrained (not based on NMR data) MD simulations correlated to the NMR data in a time-averaged ensemble. The accurate solution structure ensemble of the metal-bound protein accurately describes the role of conformational sampling in allosteric regulation of DNA binding by zinc and serves to validate our previous unrestrained MD simulations of CzrA. This methodology has potentially broad applicability in the structure determination of metal ion bound proteins, protein folding and metal template protein-design studies.






Similar content being viewed by others
Abbreviations
- AMBER:
-
Assisted model building with energy refinement
- DFT:
-
Density functional theory
- MCPB:
-
Metal center parameter builder
- MD:
-
Molecular dynamics
- MRD-NMR:
-
Metal restrained dynamics nuclear magnetic resonance
- NMR:
-
Nuclear magnetic resonance
- NOE:
-
Nuclear Overhauser enhancements
- QM/MM:
-
Quantum mechanical/molecular mechanical
- QM/MM MD:
-
Quantum mechanical/molecular mechanical molecular dynamics
- RDC:
-
Residual dipolar couplings
- RD-NMR:
-
Restrained dynamics nuclear magnetic resonance
- RMSD:
-
Root mean square deviation
- XAS:
-
X-ray absorption spectroscopy
- PDB:
-
Protein data bank
References
Allen MP, Tildesley DJ (1987) Computer simulations of liquids. Clarendon Press, Oxford
Arunkumar AI, Campanello GC, Giedroc DP (2009) Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc Natl Acad Sci USA 106(43):18177–18182. doi:10.1073/Pnas.0905558106
Azurmendi HF, Bush CA (2002) Tracking alignment from the moment of inertia tensor (TRAMITE) of biomolecules in neutral dilute liquid crystal solutions. J Am Chem Soc 124(11):2426–2427. doi:10.1031/ja017524z
Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Cavet JS, Dennison C, Graham AI, Harvie DR, Robinson NJ (2007) NMR structural analysis of cadmium sensing by winged helix repressor CmtR. J Biol Chem 282(41):30181–30188. doi:10.1074/jbc.M701119200
Bertini I, Case DA, Ferella L, Giachetti A, Rosato A (2011) A Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics 27(17):2384–2390. doi:10.1093/bioinformatics/btr415
Boehr DD, Nussinov R, Wright PE (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5(11):789–796. doi:10.1038/nchembio.232
Brockmann C, Soucek S, Kuhlmann SI, Mills-Lujan K, Kelly SM, Yang JC, Iglesias N, Stutz F, Corbett AH, Neuhaus D, Stewart M (2012) Structural basis for polyadenosine-RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export. Structure 20(6):1007–1018. doi:10.1016/j.str.2012.03.011
Brodin JD, Medina-Morales A, Ni T, Salgado EN, Ambroggio XI, Tezcan FA (2010) Evolution of metal selectivity in templated protein interfaces. J Am Chem Soc 132(25):8610–8617. doi:10.1021/ja910844n
Brunger AT (1997) X-ray crystallography and NMR reveal complementary views of structure and dynamics. Nat Struct Biol 4:862–865
Brunger AT (2007) Version 1.2 of the crystallography and NMR system. Nat Protoc 2(11):2728–2733. doi:10.1038/nprot.2007.406
Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54(Pt 5):905–921
Calhoun JR, Liu W, Spiegel K, Dal Peraro M, Klein ML, Valentine KG, Wand AJ, DeGrado WF (2008) Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement. Structure 16(2):210–215. doi:10.1016/j.str.2007.11.011
Campanello GC, Ma Z, Grossoehme NE, Guerra AJ, Ward BP, DiMarchi RD, Ye Y, Dann, CE, Giedroc DP (2013) Allosteric inhibition of a zinc-sensing transcriptional repressor: insights into the Arsenic Repressor (ArsR) Family. J Mol Biol 425:1143–1157
Case DA (2002) Molecular dynamics and NMR spin relaxation in proteins. Acc Chem Res 35(6):325–331
Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26(16):1668–1688. doi:10.1002/jcc.20290
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2010) AMBER 11. University of California, San Francisco
Chakravorty DK, Hammes-Schiffer S (2010) Impact of mutation on proton transfer reactions in ketosteroid isomerase: insights from molecular dynamics simulations. J Am Chem Soc 132(21):7549–7555. doi:10.1021/ja102714u
Chakravorty DK, Kumarasiri M, Soudackov AV, Hammes-Schiffer S (2008) Implementation of umbrella integration within the framework of the empirical valence bond approach. J Chem Theory Comput 4(11):1974–1980. doi:10.1021/ct8003386
Chakravorty DK, Soudackov AV, Hammes-Schiffer S (2009) Hybrid quantum/classical molecular dynamics simulations of the proton transfer reactions catalyzed by ketosteroid isomerase: analysis of hydrogen bonding, conformational motions, and electrostatics. Biochemistry 48(44):10608–10619. doi:10.1021/bi901353v
Chakravorty DK, Wang B, Ucisik MN, Merz KM Jr (2011) Insight into the cation-pi interaction at the metal binding site of the copper metallochaperone CusF. J Am Chem Soc 133(48):19330–19333. doi:10.1021/ja208662z
Chakravorty DK, Parker TM, Guerra AJ, Sherrill CD, Giedroc DP, Merz KM, Jr. (2012a) Energetics of zinc-mediated interactions in the allosteric pathways of metal sensor proteins. J Am Chem Soc. doi:10.1021/ja309170g
Chakravorty DK, Wang B, Lee CW, Giedroc DP, Merz KM Jr (2012b) Simulations of allosteric motions in the zinc sensor CzrA. J Am Chem Soc 134(7):3367–3376. doi:10.1021/ja208047b
Chen J, Im W, Brooks CL 3rd (2004) Refinement of NMR structures using implicit solvent and advanced sampling techniques. J Am Chem Soc 126(49):16038–16047. doi:10.1021/ja047624f
Chen J, Won HS, Im W, Dyson HJ, Brooks CL 3rd (2005) Generation of native-like protein structures from limited NMR data, modern force fields and advanced conformational sampling. J Biomol NMR 31(1):59–64. doi:10.1007/s10858-004-6056-z
Chennubhotla C, Bahar I (2006) Markov propagation of allosteric effects in biomolecular systems: application to GroEL-GroES. Mol Syst Biol 2:36. doi:10.1038/msb4100075
Clore GM, Schwieters CD (2006) Concordance of residual dipolar couplings, backbone order parameters and crystallographic B-factors for a small alpha/beta protein: a unified picture of high probability, fast atomic motions in proteins. J Mol Biol 355(5):879–886. doi:10.1016/j.jmb.2005.11.042
Cornell WD, Cieplak P, Bayly CI, Kollman PA (1993) Application of RESP charges to calculate conformational energies, hydrogen-bond energies, and free-energies of solvation. J Am Chem Soc 115(21):9620–9631
Cramer CJ, Truhlar DG (2009) Density functional theory for transition metals and transition metal chemistry. Phys Chem Chem Phys 11(46):10757–10816. doi:10.1039/B907148b
Deeth RJ, Anastasi A, Diedrich C, Randell K (2009) Molecular modelling for transition metal complexes: dealing with d-electron effects. Coordin Chem Rev 253(5–6):795–816. doi:10.1016/J.Ccr.2008.06.018
Ditchfie R, Hehre WJ, Pople JA (1971) Self-consistent molecular-orbital methods. 9. extended gaussian-type basis for molecular-orbital studies of organic molecules. J Chem Phys 54(2):724–728
Eicken C, Pennella MA, Chen XH, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP (2003) A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol 333(4):683–695. doi:10.1016/J/Jmb.2003.09.007
Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D (2005) Intrinsic dynamics of an enzyme underlies catalysis. Nature 438(7064):117–121. doi:10.1038/nature04105
Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D (2010) Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat Struct Mol Biol 17(2):241–243. doi:10.1038/nsmb.1747
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JAJ, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamao C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople J (2009) Gaussian 09. A.01 edn. Gaussian Inc., Wallingford, CT
Gao Y, Kaluarachchi K, Giedroc DP (1998) Solution structure and backbone dynamics of Mason-Pfizer monkey virus (MPMV) nucleocapsid protein. Protein Sci 7(11):2265–2280. doi:10.1002/pro.5560071104
Gaus M, Cui QA, Elstner M (2011) DFTB3: extension of the self-consistent-charge density-functional tight-binding method (SCC-DFTB). J Chem Theory Comput 7(4):931–948. doi:10.1021/Ct100684s
Giedroc DP, Arunkumar AI (2007) Metal sensor proteins: nature’s metalloregulated allosteric switches. Dalton Trans 29:3107–3120. doi:10.1039/B706769k
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A (2005) H++: a server for estimating pK(a)s and adding missing hydrogens to macromolecules. Nucleic Acids Res 33:W368–W371. doi:10.1093/Nar/Gki464
Grossoehme NE, Giedroc DP (2009) Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc 131(49):17860–17870. doi:10.1021/Ja906131b
Guntert P, Braun W, Wuthrich K (1991) Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol 217(3):517–530
Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273(1):283–298. doi:10.1006/jmbi.1997.1284
Hartsough DS, Merz KM (1995) Dynamic force-field models - molecular-dynamics simulations of human carbonic-anhydrase-II using a quantum-mechanical molecular mechanical coupled potential. J Phys Chem 99(28):11266–11275
Hay PJ, Wadt WR (1985) Abinitio effective core potentials for molecular calculations: potentials for K to Au including the outermost core orbitals. J Chem Phys 82(1):299–310
Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D (2007) Intrinsic motions along an enzymatic reaction trajectory. Nature 450(7171):838–844. doi:10.1038/nature06410
Herrmann T, Guntert P, Wuthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319(1):209–227
Hoops SC, Anderson KW, Merz KM (1991) Force-field design for metalloproteins. J Am Chem Soc 113(22):8262–8270
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65(3):712–725. doi:10.1002/prot.21123
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33
Iwahara J, Zweckstetter M, Clore GM (2006) NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci USA 103(41):15062–15067. doi:10.1073/Pnas.0605868103
Jorgensen WL, Tirado-Rives J (2005) Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci USA 102:6665–6670
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935
Joung IS, Cheatham TE 3rd (2008) Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B 112(30):9020–9041. doi:10.1021/jp8001614
Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D (2012) Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci USA 109(27):10873–10878. doi:10.1073/pnas.1203013109
Lee CW, Chakravorty DK, Chang FM, Reyes-Caballero H, Ye Y, Merz KM Jr, Giedroc DP (2012) Solution structure of Mycobacterium tuberculosis NmtR in the apo state: insights into Ni(II)-mediated allostery. Biochemistry 51(12):2619–2629. doi:10.1021/bi3001402
Li W, Zhang J, Wang J, Wang W (2008) Metal-coupled folding of Cys2His2 zinc-finger. J Am Chem Soc 130(3):892–900. doi:10.1021/ja075302g
Li X, Hayik SA, Merz KM Jr (2010) QM/MM X-ray refinement of zinc metalloenzymes. J Inorg Biochem 104(5):512–522. doi:10.1016/j.jinorgbio.2009.12.022
Lin F, Wang RX (2010) Systematic derivation of AMBER force field parameters applicable to zinc-containing systems. J Chem Theory Comput 6(6):1852–1870. doi:10.1021/Ct900454q
Long D, Bruschweiler R (2011) In silico elucidation of the recognition dynamics of ubiquitin. PLoS Comput Biol 7(4):e1002035. doi:10.1371/journal.pcbi.1002035
Lopez-Mendez B, Guntert P (2006) Automated protein structure determination from NMR spectra. J Am Chem Soc 128(40):13112–13122. doi:10.1021/ja061136l
Ma Z, Jacobsen FE, Giedroc DP (2009) Coordination chemistry of bacterial metal transport and sensing. Chem Rev 109(10):4644–4681. doi:10.1021/Cr900077w
Markwick PR, Cervantes CF, Abel BL, Komives EA, Blackledge M, McCammon JA (2010) Enhanced conformational space sampling improves the prediction of chemical shifts in proteins. J Am Chem Soc 132(4):1220–1221. doi:10.1021/ja9093692
Marsh JA, Forman-Kay JD (2009) Structure and disorder in an unfolded state under nondenaturing conditions from ensemble models consistent with a large number of experimental restraints. J Mol Biol 391(2):359–374. doi:10.1016/j.jmb.2009.06.001
Mealman TD, Zhou MW, Affandi T, Chacon KN, Aranguren ME, Blackburn NJ, Wysocki VH, McEvoy MM (2012) N-terminal region of CusB Is sufficient for metal binding and metal transfer with the metallochaperone CusF. Biochemistry 51(34):6767–6775. doi:10.1021/Bi300596a
Montalvao RW, De Simone A, Vendruscolo M (2012) Determination of structural fluctuations of proteins from structure-based calculations of residual dipolar couplings. J Biomol NMR 53(4):281–292. doi:10.1007/s10858-012-9644-3
Mumenthaler C, Guntert P, Braun W, Wuthrich K (1997) Automated combined assignment of NOESY spectra and three-dimensional protein structure determination. J Biomol NMR 10(4):351–362
Pang YP (2001) Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method. Proteins 45(3):183–189
Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP (2003) Structural elements of metal selectivity in metal sensor proteins. Proc Natl Acad Sci USA 100(7):3713–3718. doi:10.1073/Pnas.0636943100
Pennella MA, Arunkumar AI, Giedroc DP (2006) Individual metal ligands play distinct functional roles in the zinc sensor Staphylococcus aureus CzrA. J Mol Biol 356(5):1124–1136. doi:10.1016/J.Jmb.2005.12.019
Peters MB, Yang Y, Wang B, Fusti-Molnar L, Weaver MN, Merz KM (2010) Structural survey of zinc-containing proteins and development of the zinc AMBER force field (ZAFF). J Chem Theory Comput 6(9):2935–2947. doi:10.1021/Ct1002626
Pierce LCT, Salomon-Ferrer R, de Oliveira CAF, McCammon JA, Walker RC (2012) Routine access to millisecond time scale events with accelerated molecular dynamics. J Chem Theory Comput 8(9):2997–3002. doi:10.1021/Ct300284c
Ponomarev SY, Click TH, Kaminski GA (2011) Electrostatic polarization is crucial in reproducing Cu(I) interaction energies and hydration. J Phys Chem B 115(33):10079–10085. doi:10.1021/jp2051933
Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, Montelione GT, Baker D (2010) NMR structure determination for larger proteins using backbone-only data. Science 327(5968):1014–1018. doi:10.1126/science.1183649
Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA (2009) Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins 75(1):147–167. doi:10.1002/prot.22229
Rey J, Savin A (1998) Virtual space level shifting and correlation energies. Int J Quantum Chem 69(4):581–590
Roberts BP, Miller BR 3rd, Roitberg AE, Merz KM Jr (2012) Wide-open flaps are key to urease activity. J Am Chem Soc 134(24):9934–9937. doi:10.1021/ja3043239
Robustelli P, Kohlhoff K, Cavalli A, Vendruscolo M (2010) Using NMR chemical shifts as structural restraints in molecular dynamics simulations of proteins. Structure 18(8):923–933. doi:10.1016/j.str.2010.04.016
Rosato A, Aramini JM, Arrowsmith C, Bagaria A, Baker D, Cavalli A, Doreleijers JF, Eletsky A, Giachetti A, Guerry P, Gutmanas A, Guntert P, He Y, Herrmann T, Huang YJ, Jaravine V, Jonker HR, Kennedy MA, Lange OF, Liu G, Malliavin TE, Mani R, Mao B, Montelione GT, Nilges M, Rossi P, van der Schot G, Schwalbe H, Szyperski TA, Vendruscolo M, Vernon R, Vranken WF, Vries S, Vuister GW, Wu B, Yang Y, Bonvin AM (2012) Blind testing of routine, fully automated determination of protein structures from NMR data. Structure 20(2):227–236. doi:10.1016/j.str.2012.01.002
Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B, Tezcan FA (2010) Metal templated design of protein interfaces. Proc Natl Acad Sci USA 107(5):1827–1832. doi:10.1073/pnas.0906852107
Salgado EN, Brodin JD, To MM, Tezcan FA (2011) Templated construction of a Zn-selective protein dimerization motif. Inorg Chem 50(13):6323–6329. doi:10.1021/ic200746m
Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160(1):65–73
Seabra GD, Walker RC, Elstner M, Case DA, Roitberg AE (2007) Implementation of the SCC-DFTB method for hybrid QM/MM simulations within the amber molecular dynamics package. J Phys Chem A 111(26):5655–5664. doi:10.1021/Jp0700711
Senn HM, Thiel W (2009) QM/MM methods for biomolecular systems. Angew Chem Int Ed Engl 48(7):1198–1229. doi:10.1002/anie.200802019
Settergren NM, Buhlmann P, Amin EA (2008) Assessment of density functionals, semiempirical methods, and SCC-DFTB for protonated creatinine geometries. J Mol Struc-Theochem 861(1–3):68–73. doi:10.1016/J.Theochem.2008.04.010
Sgrignani J, Pierattelli R (2012) Nuclear magnetic resonance signal chemical shifts and molecular simulations: a multidisciplinary approach to modeling copper protein structures. J Biol Inorg Chem 17(1):71–79. doi:10.1007/s00775-011-0830-7
Showalter SA, Johnson E, Rance M, Bruschweiler R (2007) Toward quantitative interpretation of methyl side-chain dynamics from NMR by molecular dynamics simulations. J Am Chem Soc 129(46):14146–14147. doi:10.1021/ja075976r
Simone AD, Montalvao RW, Vendruscolo M (2011) Determination of conformational equilibria in proteins using residual dipolar couplings. J Chem Theory Comput 7(12):4189–4195. doi:101021/ct200361b
Snyder DA, Bhattacharya A, Huang YJ, Montelione GT (2005) Assessing precision and accuracy of protein structures derived from NMR data. Proteins 59(4):655–661. doi:10.1002/prot.20499
Stelzer AC, Frank AT, Bailor MH, Andricioaei I, Al-Hashimi HM (2009) Constructing atomic-resolution RNA structural ensembles using MD and motionally decoupled NMR RDCs. Methods 49:167–173. doi:10.1016/j.ymeth.2009.08.006
Stote R, Standing L (1995) Serial and multiple homicide: is there an epidemic. Soc Behav Personal 23(4):313–317
Suite (2012) Maestro, version 9.3 edn. Schrödinger, LLC, New York, NY
Thompson JM, Sgourakis NG, Liu G, Rossi P, Tang Y, Mills JL, Szyperski T, Montelione GT, Baker D (2012) Accurate protein structure modeling using sparse NMR data and homologous structure information. Proc Natl Acad Sci USA 109(25):9875–9880. doi:10.1073/pnas.1202485109
Thomson AJ, Gray HB (1998) Bio-inorganic chemistry. Curr Opin Chem Biol 2(2):155–158
Tjandra N, Bax A (1997) Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science 278(5340):1111–1114
Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH (1997) NMR evidence for slow collective motions in cyanometmyoglobin. Nat Struct Biol 4(4):292–297
Tools M-DI (2011) Maestro-desmond interoperability tools. Version 3.0 edn. Schrödinger, New York, NY
Toulouse J, Savin A, Adamo C (2002) Validation and assessment of an accurate approach to the correlation problem in density functional theory: the Kriger–Chen–Iafrate–Savin model. J Chem Phys 117(23):10465–10473. doi:10.1063/1.1521432
Tus A, Rakipovic A, Peretin G, Tomic S, Sikic M (2012) BioMe: biologically relevant metals. Nucleic Acids Res 40(Web Server issue):W352–W357. doi:10.1093/nar/gks514
Vedani A, Huhta DW (1990) A new force-field for modeling metalloproteins. J Am Chem Soc 112(12):4759–4767
Wadt WR, Hay PJ (1985) Abinitio effective core potentials for molecular calculations: potentials for main group elements Na to Bi. J Chem Phys 82(1):284–298
Waldron KJ, Robinson NJ (2009) How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol 7(1):25–35. doi:10.1038/nrmicro2057
Wang C, Vernon R, Lange O, Tyka M, Baker D (2010) Prediction of structures of zinc-binding proteins through explicit modeling of metal coordination geometry. Protein Sci 19(3):494–506. doi:10.1002/pro.327
Wu JC, Piquemal JP, Chaudret R, Reinhardt P, Ren P (2010) Polarizable molecular dynamics simulation of Zn(II) in water using the AMOEBA force field. J Chem Theory Comput 6(7):2059–2070. doi:10.1021/ct100091j
Wu RB, Lu ZY, Cao ZX, Zhang YK (2011) A transferable nonbonded pairwise force field to model zinc interactions in metalloproteins. J Chem Theory Comput 7(2):433–443. doi:10.1021/Ct100525r
Yang LW, Eyal E, Chennubhotla C, Jee J, Gronenborn AM, Bahar I (2007) Insights into equilibrium dynamics of proteins from comparison of NMR and X-ray data with computational predictions. Structure 15(6):741–749. doi:10.1016/j.str.2007.04.014
Yang Y, Chakravorty DK, Merz KM Jr (2010) Finding a needle in the haystack: computational modeling of Mg2+ binding in the active site of protein farnesyltransferase. Biochemistry 49(44):9658–9666. doi:10.1021/bi1008358
Yang Y, Miao Y, Wang B, Cui G, Merz KM Jr (2012a) Catalytic mechanism of aromatic prenylation by NphB. Biochemistry 51(12):2606–2618. doi:10.1021/bi201800m
Yang Y, Wang B, Ucisik MN, Cui G, Fierke CA, Merz KM Jr (2012b) Insights into the mechanistic dichotomy of the protein farnesyltransferase peptide substrates CVIM and CVLS. J Am Chem Soc 134(2):820–823. doi:10.1021/ja209650h
York DM, Darden TA, Pedersen LG (1993) The effect of long-range electrostatic interactions in simulations of macromolecular crystals: a comparison of the Ewald and truncated list methods. J Chem Phys 99(10):8345–8348
Zhao Y, Truhlar DG (2008) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Acc 120(1–3):215–241. doi:10.1007/S00214-007-0310-X
Zweckstetter M, Bax A (2000) Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J Am Chem Soc 122(15):3791–3792
Acknowledgments
We gratefully acknowledge the United States National Institutes of Health for support of this study (GM044974 to K.M.M. and GM042569 to D.P.G.) and high performance computing at the University of Florida (UFHPC) for providing and maintaining the computational resources used to perform this study. We also thank Sarah Gordon for helpful discussions.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chakravorty, D.K., Wang, B., Lee, C.W. et al. Solution NMR refinement of a metal ion bound protein using metal ion inclusive restrained molecular dynamics methods. J Biomol NMR 56, 125–137 (2013). https://doi.org/10.1007/s10858-013-9729-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9729-7