Abstract
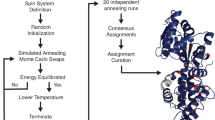
We describe a general computational approach to site-specific resonance assignments in multidimensional NMR studies of uniformly 15N,13C-labeled biopolymers, based on a simple Monte Carlo/simulated annealing (MCSA) algorithm contained in the program MCASSIGN2. Input to MCASSIGN2 includes lists of multidimensional signals in the NMR spectra with their possible residue-type assignments (which need not be unique), the biopolymer sequence, and a table that describes the connections that relate one signal list to another. As output, MCASSIGN2 produces a high-scoring sequential assignment of the multidimensional signals, using a score function that rewards good connections (i.e., agreement between relevant sets of chemical shifts in different signal lists) and penalizes bad connections, unassigned signals, and assignment gaps. Examination of a set of high-scoring assignments from a large number of independent runs allows one to determine whether a unique assignment exists for the entire sequence or parts thereof. We demonstrate the MCSA algorithm using two-dimensional (2D) and three-dimensional (3D) solid state NMR spectra of several model protein samples (α-spectrin SH3 domain and protein G/B1 microcrystals, HET-s218–289 fibrils), obtained with magic-angle spinning and standard polarization transfer techniques. The MCSA algorithm and MCASSIGN2 program can accommodate arbitrary combinations of NMR spectra with arbitrary dimensionality, and can therefore be applied in many areas of solid state and solution NMR.





Similar content being viewed by others
References
Bailey-Kellogg C, Widge A, Kelley JJ, Berardi MJ, Bushweller JH, Donald BR (2000) The NOESY jigsaw: automated protein secondary structure and main-chain assignment from sparse, unassigned NMR data. J Comput Biol 7:537–558
Bartels C, Guntert P, Billeter M, Wuthrich K (1997) Garant: a general algorithm for resonance assignment of multidimensional nuclear magnetic resonance spectra. J Comput Chem 18:139–149
Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG (1995) Heteronuclear decoupling in rotating solids. J Chem Phys 103:6951–6958
Bennett AE, Rienstra CM, Griffiths JM, Zhen WG, Lansbury PT, Griffin RG (1998) Homonuclear radio frequency-driven recoupling in rotating solids. J Chem Phys 108:9463–9479
Buchler NEG, Zuiderweg ERP, Wang H, Goldstein RA (1997) Protein heteronuclear NMR assignments using mean-field simulated annealing. J Magn Reson 125:34–42
Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG (2010) Magic-angle spinning NMR analysis of β2-microglobulin amyloid fibrils in two distinct morphologies. J Am Chem Soc 132:10414–10423
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRpipe: a multidimensional spectral processing system based on Unix pipes. J Biomol NMR 6:277–293
Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM (2005) Magic-angle spinning solid state NMR spectroscopy of the β1 immunoglobulin binding domain of protein G (GB1): 15N and 13 chemical shift assignments and conformational analysis. J Am Chem Soc 127:12291–12305
Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP (2008) Molecular conformation and dynamics of the Y145Stop variant of human prion protein. Proc Natl Acad Sci USA 105:6284–6289
Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP (2010) Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid state nuclear magnetic resonance spectroscopy. J Am Chem Soc 132:2393–2403
Hitchens TK, Lukin JA, Zhan YP, McCallum SA, Rule GS (2003) Monte: an automated Monte Carlo based approach to nuclear magnetic resonance assignment of proteins. J Biomol NMR 25:1–9
Hyberts SG, Wagner G (2003) IBIS: a tool for automated sequential assignment of protein spectra from triple resonance experiments. J Biomol NMR 26:335–344
Igumenova TI, Wand AJ, McDermott AE (2004) Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc 126:5323–5331
Ishii Y (2001) 13C–13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J Chem Phys 114:8473–8483
Ishii Y, Tycko R (2000) Multidimensional heteronuclear correlation spectroscopy of a uniformly 15N- and 13C-labeled peptide crystal: toward spectral resolution, assignment, and structure determination of oriented molecules in solid state NMR. J Am Chem Soc 122:1443–1455
Lemak A, Steren CA, Arrowsmith CH, Llinas M (2008) Sequence specific resonance assignment via multicanonical Monte Carlo search using an Abacus approach. J Biomol NMR 41:29–41
Leutner M, Gschwind RM, Liermann J, Schwarz C, Gemmecker G, Kessler H (1998) Automated backbone assignment of labeled proteins using the threshold accepting algorithm. J Biomol NMR 11:31–43
Li KB, Sanctuary BC (1997) Automated resonance assignment of proteins using heteronuclear 3D NMR. 2. Side chain and sequence-specific assignment. J Chem Inf Comput Sci 37:467–477
Lukin JA, Gove AP, Talukdar SN, Ho C (1997) Automated probabilistic method for assigning backbone resonances of 13C, 15N-labeled proteins. J Biomol NMR 9:151–166
McDermott A, Polenova T, Bockmann A, Zilm KW, Paulsen EK, Martin RW, Montelione GT (2000) Partial NMR assignments for uniformly 13C, 15N-enriched BPTI in the solid state. J Biomol NMR 16:209–219
Morcombe CR, Gaponenko V, Byrd RA, Zilm KW (2004) Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc 126:7196–7197
Moseley HNB, Monleon D, Montelione GT (2001) Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol 339:91–108
Moseley HNB, Sperling LJ, Rienstra CM (2010) Automated protein resonance assignments of magic angle spinning solid state NMR spectra of β1 immunoglobulin binding domain of protein G (GB1). J Biomol NMR 48:123–128
Nelson SJ, Schneider DM, Wand AJ (1991) Implementation of the main chain directed assignment strategy: computer-assisted approach. Biophys J 59:1113–1122
Pauli J, van Rossum B, Forster H, de Groot HJM, Oschkinat H (2000) Sample optimization and identification of signal patterns of amino acid side chains in 2D RFDR spectra of the α-spectrin SH3 domain. J Magn Reson 143:411–416
Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H (2001) Backbone and side-chain 13C and 15N signal assignments of the α-spectrin SH3 domain by magic-angle spinning solid state NMR at 17.6 Tesla. ChemBioChem 2:272–281
Petkova AT, Baldus M, Belenky M, Hong M, Griffin RG, Herzfeld J (2003) Backbone and side chain assignment strategies for multiply labeled membrane peptides and proteins in the solid state. J Magn Reson 160:1–12
Schmidt HLF, Sperling LJ, Gao YG, Wylie BJ, Boettcher JM, Wilson SR, Rienstra CA (2007) Crystal polymorphism of protein GB1 examined by solid state NMR spectroscopy and x-ray diffraction. J Phys Chem B 111:14362–14369
Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH (2006a) Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc 128:13224–13228
Siemer AB, Ritter C, Steinmetz MO, Ernst M, Riek R, Meier BH (2006b) 13C, 15N resonance assignment of parts of the HET-s prion protein in its amyloid form. J Biomol NMR 34:75–87
Sinha N, Grant CV, Park SH, Brown JM, Opella SJ (2007) Triple resonance experiments for aligned sample solid state NMR of 13C and 15N labeled proteins. J Magn Reson 186:51–64
Takegoshi K, Nakamura S, Terao T (2001) 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett 344:631–637
Tycko R, Hu KN (2010) A Monte Carlo/simulated annealing algorithm for sequential resonance assignment in solid state NMR of uniformly labeled proteins with magic-angle spinning. J Magn Reson 205:304–314
Tycko R, Savtchenko R, Ostapchenko VG, Makarava N, Baskakov IV (2010) The α-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel β-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry 49:9488–9497
Van Melckebeke H, Wasmer C, Lange A, Eiso AB, Loquet A, Böckmann A, Meier BH (2010) Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid state NMR spectroscopy. J Am Chem Soc 132:13765–13775
Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH (2008) Amyloid fibrils of the HET-s(218–289) prion form a β-solenoid with a triangular hydrophobic core. Science 319:1523–1526
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases within the National Institutes of Health. Computational facilities of the Helix Systems at the National Institutes of Health were used in this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, KN., Qiang, W. & Tycko, R. A general Monte Carlo/simulated annealing algorithm for resonance assignment in NMR of uniformly labeled biopolymers. J Biomol NMR 50, 267–276 (2011). https://doi.org/10.1007/s10858-011-9517-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-011-9517-1