Abstract
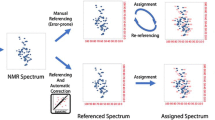
It has been estimated that more than 20% of the proteins in the BMRB are improperly referenced and that about 1% of all chemical shift assignments are mis-assigned. These statistics also reflect the likelihood that any newly assigned protein will have shift assignment or shift referencing errors. The relatively high frequency of these errors continues to be a concern for the biomolecular NMR community. While several programs do exist to detect and/or correct chemical shift mis-referencing or chemical shift mis-assignments, most can only do one, or the other. The one program (SHIFTCOR) that is capable of handling both chemical shift mis-referencing and mis-assignments, requires the 3D structure coordinates of the target protein. Given that chemical shift mis-assignments and chemical shift re-referencing issues should ideally be addressed prior to 3D structure determination, there is a clear need to develop a structure-independent approach. Here, we present a new structure-independent protocol, which is based on using residue-specific and secondary structure-specific chemical shift distributions calculated over small (3–6 residue) fragments to identify mis-assigned resonances. The method is also able to identify and re-reference mis-referenced chemical shift assignments. Comparisons against existing re-referencing or mis-assignment detection programs show that the method is as good or superior to existing approaches. The protocol described here has been implemented into a freely available Java program called “Probabilistic Approach for protein Nmr Assignment Validation (PANAV)” and as a web server (http://redpoll.pharmacy.ualberta.ca/PANAV) which can be used to validate and/or correct as well as re-reference assigned protein chemical shifts.





Similar content being viewed by others
References
de Dios AC, Pearson JG, Oldfield E (1993) Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science 260:1491–1496
Ginzinger SW, Gerick F, Coles M, Heun V (2007) CheckShift: automatic correction of inconsistent chemical shift referencing. J Biomol NMR 39:223–227
Gronenborn AM, Clore GM (1994) Identification of N-terminal helix capping boxes by means of 13C chemical shifts. J Biomol NMR 4:455–458
Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M (2009) Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J Am Chem Soc 131:13894–13895
Metzler WJ, Constantine KL, Friedrichs MS, Bell AJ, Ernst EG, Lavoie TB, Mueller L (1993) Characterization of the three-dimensional solution structure of human profiling: 1H, 13C and 15 N NMR assignments and global folding pattern. Biochemistry 32:13818–13829
Moseley HN, Sahota G, Montelione GT (2004) Assigment validation software suite for the evaluation and presentatio of protein resonance assignment data. J Biomol NMR 28:341–355
Neal S, Nip AM, Zhang H, Wishart DS (2003) Rapid and accurate calculation of protein 1H, 13C and 15 N chemical shifts. J Biomol NMR 26:215–240
Seavey BR, Farr EA, Westler WM, Markley JL (1991) A relational database for sequence-specific protein NMR data. J Biomol NMR 1:217–236
Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Sugnarapu KK, Ignatchenko A, Arrowsmith CH, Syperski T, Montelione GT, Baker D, Bax A (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA 105:4685–4690
Spera S, Bax A (1991) Empirical correlation between protein backbone conformatio and Ca and Cb 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc 113:5490–5492
Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao H, Markley JL (2008) BioMagResBank. Nucleic Acids Res 36:D402–D408
Wang Y, Jardetzky O (2002a) Investigation of the neighboring residue effects on protein chemical shifts. J Am Chem Soc 124:14075–14084
Wang Y, Jardetzky O (2002b) Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci 11:852–861
Wang Y, Jardetzky O (2004) Predicting 15 N chemical shifts in proteins using the preceding residue-specific individual shielding surfaces from phi, psi i-1 and chi 1 torsion angles. J Biomol NMR 28:327–340
Wang L, Markley JL (2009) Empirical correlation between protein backbone 15 N and 13C secondary chemical shifts and its application to nitrogen chemical shift re-referencing. J Biomol NMR 44:95–99
Wang Y, Wishart DS (2005) A simple method to adjust inconsistently referenced 13C and 15 N chemical shift assignments of proteins. J Biomol NMR 31:143–148
Wishart DS, Nip AM (1998) Protein chemical shift analysis: a practical guide. Biochem Cell Biol 76:153–163
Wishart DS, Sykes BD (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:171–180
Wishart DS, Sykes BD, Richards FM (1991) Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol 222:311–333
Wishart DS, Sykes BD, Richards FM (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31:1647–1651
Xu XP, Case DA (2001) Automated prediction of 15 N, 13Callpha, 13Cbeta and 13C’ chemical shifts in proteins using a density functional database. J Biomol NMR 21:321–333
Zhang H, Neal S, Wishart DS (2003) RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR 25:173–195
Acknowledgments
DSW wishes to acknowledge the financial support of the Natural Sciences and Engineering Research Council (NSERC) and the Alberta Prion Research Institute (APRI).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, B., Wang, Y. & Wishart, D.S. A probabilistic approach for validating protein NMR chemical shift assignments. J Biomol NMR 47, 85–99 (2010). https://doi.org/10.1007/s10858-010-9407-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-010-9407-y