Abstract
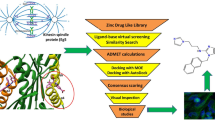
Kinases are one of the major players in cancer development and progression. Serine threonine kinases such as human checkpoint kinase-1 (Chk1), Mek1 and cyclin-dependent kinases have been identified as promising targets for cancer treatment. Chk1 is an important kinase with vital role in cell cycle arrest and many potent inhibitors targeted to Chk1 have been reported and few are currently in clinical trials. Considering the emerging importance of Chk1 inhibitors in cancer treatment there is a need to widen the chemical space of Chk1 inhibitors. In this study, we are reporting an integrated in silico approach to identify novel competitive Chk1 inhibitors. A 4-features pharmacophore model was derived from a co-crystallized structure of known potent Chk1 inhibitor and subjected to screen Maybridge compound library. Hits obtained from the screening were docked into the Chk1 active site and filtered on the basis of docking score and the number of pharmacophoric features showing conserved interaction within the active site of Chk1. Further, five compounds from the top ranking hits were subjected to in vitro evaluation as Chk1 inhibitor. After the kinase assay, four compounds were found to be active against human Chk1 (IC50 range from 4.2 to 12.5 µM). Subsequent study using the cdc25-22 mutant yeast cells revealed that one of compound (SPB07479; IC50 = 4.24 µM) promoted the formation of multinucleated cells, therefore overriding the cell cycle checkpoint. Validation studies using normal and human cancer cell lines, indicated that SPB07479 significantly inhibited proliferation of cervical cancer cells as a single agent and chemosensitized glioma and pancreatic cancer cell lines to standard chemotherapy while sparing normal cells. Additionally SPB07479 did not show significant cytotoxicity in normal cells. In conclusion we report that SPB07479 appear promising for further development of Chk1 inhibitors. This study also highlights the role of conserved water molecules in the active site of Chk1 for the successful identification of novel inhibitors.




Similar content being viewed by others
References
http://www.cancer.gov/. Accessed Dec 2013
Bogenrieder T, Herlyn M (2003) Axis of evil: molecular mechanisms of cancer metastasis. Oncogene 22(42):6524–6536
Galluzzi L, Kepp O, Heiden MG, Kroemer G (2013) Metabolic targets for cancer therapy. Nat Rev Drug Discov 12(11):829–846
Tsatsanis C, Spandidos DA (2000) The role of oncogenic kinases in human cancer (review). Int J Mol Med 5(6):583–590
Fabbro D, García-Echeverría C (2002) Targeting protein kinases in cancer therapy. Curr Opin Drug Discov Dev 5(5):701–712
Hartmann JT, Haap M, Kopp HG, Lipp HP (2009) Tyrosine kinase inhibitors—a review on pharmacology, metabolism and side effects. Curr Drug Metab 10(5):470–481
Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M et al (2006) Frequent alterations in the expression of serine/threonine kinases in human cancers. Cancer Res 66(16):8147–8154
Tao ZF, Lin NH (2006) Chk1 inhibitors for novel cancer treatment. Anticancer Agents. Med Chem 6(4):377–388
Chang-Yew Leow C, Gerondakis S, Spencer A (2013) MEK inhibitors as a chemotherapeutic intervention in multiple myeloma. Blood Cancer J 3:e105. doi:10.1038/bcj.2013.1
Merkel AL, Meggers E, Ocker M (2012) PIM1 kinase as a target for cancer therapy. Expert Opin Investig Drugs 21(4):425–436
Malumbres M, Barbacid M (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9(3):153–166
Chen P, Luo C, Deng Y, Ryan K, Register J et al (2000) The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell 100(6):681–692
Dent P, Tang Y, Yacoub A, Dai Y, Fisher PB, Grant S (2011) CHK1 inhibitors incombination chemotherapy: thinking beyond the cell cycle. Mol Interv 11(2):133–140
Adams JL, Lee D (1999) Recent progress towards the identification of selective inhibitors of serine/threonine protein kinases. Curr Opin Drug Discov Dev 2(2):96–109
Walton MI, Eve PD, Hayes A, Valenti MR, De Haven Brandon AK, Box G et al (2012) CCT244747 is a novel potent and selective CHK1 inhibitor with oral efficacy alone and in combination with genotoxic anticancer drugs. Clin Cancer Res 18(20):5650–5661
Reader JC, Matthews TP, Klair S, Cheung KM, Scanlon J, Proisy N et al (2011) Structure-guided evolution of potent and selective CHK1 inhibitors through scaffold morphing. J Med Chem 54(24):8328–8342
Li T, Christensen SD, Frankel PH, Margolin KA, Agarwala SS et al (2012) A phase II study of cell cycle inhibitor UCN-01 in patients with metastatic melanoma: a California cancer consortium trial. Investig New Drugs 30(2):741–748
Vanderpool D, Johnson TO, Ping C, Bergqvist S, Alton G et al (2009) Characterization of the CHK1 allosteric inhibitor binding site. Biochemistry 48(41):9823–9830
Converso A, Hartingh T, Garbaccio RM, Tasber E, Rickert K et al (2009) Development of thioquinazolinones, allosteric Chk1 kinase inhibitors. Bioorg Med Chem Lett 19(4):1240–1244
Chen JJ, Liu TL, Yang LJ, Li LL, Wei YQ, Yang SY (2009) Pharmacophore modeling and virtual screening studies of checkpoint kinase 1 inhibitors. Chem Pharm Bull (Tokyo) 57(7):704–709
Chen XM, Lu T, Lu S, Li HF, Yuan HL et al (2010) Structure-based and shape-complemented pharmacophore modeling for the discovery of novel checkpoint kinase 1 inhibitors. J Mol Model 16(7):1195–1204
Ashwell S, Thomas G, Ioannidis S, Janetka J, Lyne P, Su M, Toader D, Yu Y (2005) WO2005066163. Accessed Dec 2013
Zhao L, Zhang Y, Dai C, Guzi T, Wiswell D et al (2010) Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors. Bioorg Med Chem Lett 20(24):7216–7221
SYBYL7.1 Tripos Inc., St. Louis, MO, 63144, USA
Hurst T (1994) Flexible 3D searching: the directed tweak technique. J Chem Inf Comput Sci 34:190–196
Kramer B, Rarey M, Lengauer T (1999) Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins 37(2):228–241
Schellhammer I, Rarey M (2004) FlexX-Scan: fast, structure-based virtual screening. Proteins 57(3):504–517
Foloppe N, Fisher LM, Francis G, Howes R, Kierstan P, Potter A (2006) Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification. Bioorg Med Chem 14(6):1792–1804
Tao ZF, Wang L, Stewart KD, Chen Z, Gu W et al (2007) Structure-based design, synthesis, and biological evaluation of potent and selective macrocyclic checkpoint kinase 1 inhibitors. J Med Chem 50(7):1514–1527
Kuntz ID, Chen K, Sharp KA, Kollman PA (1999) The maximal affinity of ligands. Proc Natl Acad Sci USA 96(18):9997–10002
Hopkins AL, Groom CR, Alex A (2004) Ligand efficiency: a useful metric for lead selection. Drug Discov Today 9(10):430–431
Hopkins AL, Keserü GM, Leeson PD, Rees DC, Reynolds CH (2014) The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov 13(2):105–121
Abad-Zapatero C (2007) Ligand efficiency indices for effective drug discovery. Expert Opin Drug Discov 2(4):469–488
Symyx Software: MACCS structural keys San Ramon, CA
McGovern SL, Shoichet BK (2003) Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J Med Chem 46(14):2895–2907
Walworth N, Davey S, Beach D (1993) Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature (Lond.) 363:368–371
Carr AM, Moudjou M, Bentley NJ, Hagan IM (1995) The chk1 pathway is required to prevent mitosis following cell-cycle arrest at ‘start’. Curr Biol 5:1179–1190
Kumagai A, Guo Z, Emami KH, Wang S, Dunphy WG (1998) The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol 142:1559–1569
Furnari B, Blasina A, Boddy M, McGowan C, Russell P (1999) Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol Biol Cell 10:833–845
Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD et al (2009) Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther 8(1):45–54
Tang Y, Dai Y, Grant S, Dent P (2012) Enhancing CHK1 inhibitor lethality in glioblastoma. Cancer Biol Ther 13(6):379–388
Keseru GM, Makara GM (2006) Hit discovery and hit-to-lead approaches. Drug Discov Today 11(15–16):741–748
Daud A, Springett GM, Mendelson DS, Munster PN, Goldman JR et al. (2010) A Phase I dose-escalation study of SCH 900776, a selective inhibitor of checkpoint kinase 1 (CHK1), in combination with gemcitabine (Gem) in subjects with advanced solid tumors. J Clin Oncol 28 (Suppl): Abstract 3064
McNeely SC, Burke TF, DurlandBusbice S, Barnard DS, Marshall MS, Bence AK, Beckmann RP (2011). LY2606368, a second generation Chk1 inhibitor, inhibits growth of ovarian carcinoma xenografts either as monotherapy or in combination with standard-of-care agents. AACR-NCI-EORTC International Conference: Mol Cancer Ther 10 (Suppl 1): Abstract A108
Wu W, Bi C, Bence AK, Um SL, Yan B, Starling JJ, Marshall MS, Beckmann RP (2012) Antitumor activity of Chk1 inhibitor LY2606368 as a single agent in SW1990 human pancreas orthotopic tumor model. AACR 103rd Annual Meeting2012. Cancer Res 72 (Suppl 1): Abstract 1776
Walton MI, Eve PD, Hayes A, Valenti M, De Haven Brandon A et al (2010) The preclinical pharmacology and therapeutic activity of the novel CHK1 inhibitor SAR-020106. Mol Cancer Ther 9:89–100
Foloppe N, Fisher LM, Howes R, Potter A, Robertson AG, Surgenor AE (2006) Identification of chemically diverse Chk1 inhibitors by receptor-based virtual screening. Bioorg Med Chem 14(14):4792–4802
Matthews TP, McHardy T, Klair S, Boxall K, Fisher M et al (2010) Design and evaluation of 3,6-di(hetero)aryl imidazo[1,2-a]pyrazines as inhibitors of checkpoint and other kinases. Bioorg Med Chem Lett 20(14):4045–4049
Dudkin VY, Rickert K, Kreatsoulas C, Wang C, Arrington KL et al (2012) Pyridyl aminothiazoles as potent inhibitors of Chk1 with slow dissociation rates. Bioorg Med Chem Lett 22(7):2609–2612
Acknowledgments
Work reported in this project is supported by grants from CSIR network projects GENESIS (BSC0121) and UNDO (BSC0103). VK and SK acknowledge DBT and ICMR for their fellowships respectively. This manuscript is a CSIR-CDRI communication number 8815.
Author information
Authors and Affiliations
Corresponding author
Additional information
Vikash Kumar and Saman Khan have contributed equally to this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, V., Khan, S., Gupta, P. et al. Identification of novel inhibitors of human Chk1 using pharmacophore-based virtual screening and their evaluation as potential anti-cancer agents. J Comput Aided Mol Des 28, 1247–1256 (2014). https://doi.org/10.1007/s10822-014-9800-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9800-9