Abstract
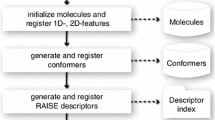
Structure-based virtual screening today is basically organized as a sequential process where the molecules of a screening library are evaluated for instance with respect to their fit with a biological target. In this paper, we present a novel structure-based screening paradigm avoiding sequential searching and therefore enabling sublinear runtime behavior. We implemented the novel paradigm in the virtual screening tool TrixX and successfully applied it in screening experiments on four targets from relevant therapeutic areas. With the screening paradigm implemented in TrixX, we propose some important extensions and modifications to traditional virtual screening approaches: Instead of processing all compounds in the screening library sequentially, TrixX first analyzes the geometric and physicochemical binding site characteristics and then draws compounds with matching features from a compound catalog. The catalog organizes the compounds by their physicochemical and geometric features making use of relational database technology with indexed tables in order to support efficient queries for compounds with specific features. A key element of the compound catalog is a highly selective geometric descriptor that carries information on the type of functional groups of the compound, their Euclidian distance, the preferred interaction direction of each functional group, and the location of steric bulk around the triangle.
In a re-docking experiment with 200 protein–ligand complexes, we could show that TrixX is able to correctly predict the location of ligand functional groups in co-crystallized complexes. In a retrospective virtual screening experiment for four different targets, the enrichment factors of TrixX are comparable to the enrichment factors of FlexX and FlexX-Scan. With computing times clearly below one second per compound, TrixX counts among the fastest virtual screening tools currently available and is nearly two orders of magnitude faster than standard FlexX.











Similar content being viewed by others
Notes
This runtime behavior is valid within the tested library sizes. Libraries with more than 130,000 compounds demand more main memory and may require even more selective molecule descriptors. Further, experiments need to prove or disprove whether the observed runtime behavior can be further extrapolated.
Hardware environment A: 32-bit version of TrixX on a 2.4 GHz Dual Xeon workstation with 4 GB of main memory. Hardware environment B: 64-bit version of TrixX on a SUN Fire server using a one of four CPUs and 32 GB of main memory: We observed similar average runtimes of TrixX on both hardware environments.
The gap of re-docking performance between TrixX and FlexX/FlexX-Scan (number of correct predictions) is rather large for very accurate solutions (≤1.0 Å) and tends to close for less accurate solutions (≤2.5 Å).
References
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Nat Rev Drug Discov 3(11):935
Bajorath J (2002) Nat Rev Drug Discov 1(11):882
Wang R, Lu Y, Wang S (2003) J Med Chem 46(12):2287
Wang R, Lu Y, Fang X, Wang S (2004) J Chem Inf Comput Sci 44(6):2114
Stockwell BR (2004) Nature 432(7019):846
Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE (1982) J Mol Biol 161(2):269
Hindle SA, Rarey M, Buning C, Lengauer T (2002) J Comput Aided Mol Des 16(2):129
Good AC, Krystek SR, Mason JS (2000) Drug Discov Today 5(12):S61
Claussen H, Gastreich M, Apelt V, Greene J, Hindle SA, Lemmen C (2004) Curr Drug Discov Technol 1:49
Zuccotto F (2003) J Chem Inf Comput Sci 43(5):1542
Renner S, Schneider G (2004) J Med Chem 47(19):4653
Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Arthurs S, Colson AB, Freer ST, Larson V, Luty BA, Marrone T, Rose PW (2000) J Comput Aided Mol Des 14:731
Kraemer A, Horn HW, Rice JE (2003) J Comput Aided Mol Des 17:13
Mason JS, Morize I, Menard PR, Cheney DL, Hulme C, Labaudiniere RF (1999) J Med Chem 42:3251
Schellhammer I, Rarey M (2004) Proteins 57(3):504
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) J Med Chem 47(7):1739
Floriano WB, Vaidehi N, Zamanakos G, Goddard III WA (2004) J Med Chem 47(1):56
Rarey M, Lengauer T (2000) Persp Drug Discov Des 20:63
Sun Y, Ewing TJ, Skillman AG, Kuntz ID (1998) J Comput Aided Mol Des 12(6):597
Lorber DM, Shoichet BK (1998) Protein Sci 7:938
Su AI, Lorber DM, Weston GS, Baase WA, Matthews BW, Shoichet BK (2001) Proteins 42(2):279
Joseph-McCarthy D, Thomas IV BE, Belmarsh M, Moustakas D, Alvarez JC (2003) Proteins 51:172
Schnecke V, Swanson CA, Getzoff ED, Tainer JA, Kuhn LA (1998) Proteins 33(1):74
Schnecke V, Kuhn LA (2000) Persp Drug Discov Des 20:171
Weininger D, Weininger A, Weininger JL (1989) J Chem Inf Comput Sci 29:97
Weininger D (1988) J Chem Inf Comput Sci 28(1):31
Bayer R, McCreight E (1972) Acta Informatica 1(3):173
Kramer B, Rarey M, Lengauer T (1999) Proteins 37:228
Stahl M, Rarey M (2001) J Med Chem 44:1035
Wildman SA, Crippen GM (1999) J Chem Inf Comput Sci 39:868
Banner DW, Hadvary P (1991) J Biol Chem 266(30):20085
Böhm HJ Thrombin-Inhibitors, collected experimental data, personal communication
Bolin JT, Filman DJ, Matthews DA, Hamlin RC, Kraut J (1982) J Biol Chem 257(22):13650
Selassie CD, Fang ZX, Li RL, Hansch C, Debnath G, Klein TE, Langridge R, Kaufman BT (1989) J Med Chem 32(8):1895
Furet P, Meyer T, Strauss A, Raccuglia S, Rondeau JM (2002) Bioorg Med Chem Lett 12(2):221
Natesh R, Schwager SL, Sturrock ED, Acharya KR (2003) Nature 421(6922):551
Acknowledgment
The authors thank BioSolveIT GmbH (St. Augustin, Germany) and AstraZeneca (Mölndal, Sweden) for funding our work. We are grateful for constructive discussions on the molecule descriptor and on method validation with colleagues from AstraZeneca and BioSolveIT, especially Jens Sadowski and Christian Lemmen.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schellhammer, I., Rarey, M. TrixX: structure-based molecule indexing for large-scale virtual screening in sublinear time. J Comput Aided Mol Des 21, 223–238 (2007). https://doi.org/10.1007/s10822-007-9103-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-007-9103-5