Abstract
Transaction logs from online search engines are valuable for two reasons: First, they provide insight into human information-seeking behavior. Second, log data can be used to train user models, which can then be applied to improve retrieval systems. This article presents a study of logs from PubMed®, the public gateway to the MEDLINE® database of bibliographic records from the medical and biomedical primary literature. Unlike most previous studies on general Web search, our work examines user activities with a highly-specialized search engine. We encode user actions as string sequences and model these sequences using n-gram language models. The models are evaluated in terms of perplexity and in a sequence prediction task. They help us better understand how PubMed users search for information and provide an enabler for improving users’ search experience.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Information seeking is fundamentally an iterative activity that involves multiple interactions between a user and a search system. With the advent of online search engines, it has become standard practice to store records of users’ activities—commonly known as search transaction logs (or query logs, as a special case). These resources are useful for two main reasons: First, they shed light on human information-seeking behavior—what users want and how they go about accomplishing it (e.g., Silverstein et al. 1999; Beitzel et al. 2004; Rose and Levinson 2004; Jansen and Spink 2006). Second, log data can be exploited to improve the user’s search experience; examples include query suggestion (Anick 2003; Cui et al. 2003) and improved ranking (Agichtein et al. 2006; Joachims et al. 2007).
The nature of users’ interactions with search engines depends on many factors, including characteristics of the user, type of task, problem domain, etc. While there has been a significant amount of work on analysis of search logs, previous studies have almost exclusively focused on general Web search; see Jansen and Spink (2006) for an overview. In this article, we present an analysis of logs from the PubMed search engine (more details in Sect. 2). Note that due to the subject domain of PubMed (the life sciences) and the demographic profile of its users (biologists, physicians, clinical researchers, etc.), our dataset differs significantly from typical Web search logs, such as those collected by Google or Yahoo, which represent a broader demographic and a greater variety of user needs. However, these differences are not the subject of this article.
Our work has two main goals: First, we wish to better understand how users interact with PubMed (Sects. 3, 5). Second, we wish to build computational models of user–system interactions as a first step to improving the search experience. This article explores the idea that users’ actions can be encoded as a string sequence and modeled using n-gram language models (Sect. 4). We evaluate these models both in terms of perplexity and in a sequence prediction task.
2 PubMed transaction logs
PubMed is the public gateway to MEDLINE, the authoritative repository of bibliographic records from the medical and biomedical primary literature, and more broadly, topics in the life sciences ranging from biophysics to public health.Footnote 1 Both resources are maintained by the U.S. National Library of Medicine (NLM). As of March 2008, MEDLINE contains over 17.8 m records (called citations) dating back to 1949, all with basic bibliographic information. In addition, most records (especially those added in more recent decades) contain abstract text, and increasingly, links to full text. Citations are added to MEDLINE at a rate of approximately 65k records a month. PubMed is frequently used by physicians, scientists (e.g., biologists, biochemists), and lay-people to solve complex tasks (particularly the first two groups). Although PubMed provides access to a wealth of resources (e.g., gene databases, sequence information, etc.), this study focuses specifically on users’ interaction with MEDLINE.
PubMed is a sophisticated Boolean search engine that allows users to query not only on title and abstract text, but also on metadata fields (e.g., journal or author) and using controlled vocabulary MeSH® terms. PubMed also provides a number of pre-defined “search templates”, which allow users to narrow the scope of the articles that are retrieved (Haynes et al. 1994). These filters are implemented as fixed Boolean query fragments that the system automatically appends to each query.
This work examines transaction logs gathered over an 8-day span, June 20–27, 2007. The basic unit of analysis is the session, which is tracked through a browser cookie. Sessions are comprised of transactions, each of which corresponds to a CGI invocation. Due to the nature of this tracking mechanism, a user who engages PubMed with multiple browser windows (or tabs) will show up as a single session, since there is no effective way to separate the source of the CGI requests. Note that our definition of a session is very coarse-grained; we explore different segmentation techniques in Sect. 3.
The logs contain a wealth of information, including timestamp and details of the CGI invocation (encoded parameters), which allows us to reconstruct with reasonable accuracy the actions of a particular user. Certain client-side actions, such as use of the browser “back” button, are not captured, although it is possible to infer some of these behaviors. This data provides a unique glimpse into the actions of PubMed users—although PubMed queries have been previously studied (Herskovic et al. 2007), this work represents the first systematic analysis of session data. Although there have been previous studies of transaction logs from search systems for the medical literature, e.g., studies on PaperChase (Horowitz et al. 1983; King 1991) and Grateful Med (Cahan 1989), our work involves significantly more data and users. It is our intention to make an appropriately-processed version of this dataset available to the research community.
In addition to the domain, we note another unique characteristic of our logs. Whereas most datasets studied by academic researchers contain only search queries (Jansen and Spink 2006), our logs capture all interactions between the user and PubMed. In fact, search queries account for only 33% of user actions. A more complete record allows us to construct richer models of users’ activities and understand their browsing behaviors (see Sect. 5).
Over the 8-day span, we observed a total of 8.68 m sessions, totaling 41.8 m transactions (for clarity, we refer to this as the raw dataset). A number of filters were first applied to discard sessions not of interest. We found that 63.2%, or 5.49 m sessions, consisted of only one transaction—most of these represent direct access into MEDLINE, e.g., through an embedded link or another search engine; this number is consistent with previous studies on Web search (Jansen and Spink 2006). Although single-transaction sessions account for a large portion of all traffic, we discarded them since they do not represent interesting user behavior. At the other end of the spectrum, we discarded all sessions with more than 500 transactions (an arbitrary threshold), since they were likely to be robots spidering the collection. This removes only 162 sessions, but 271k transactions. Finally, we removed sessions in which the user was not primarily engaged with MEDLINE. In addition to bibliographic records, PubMed provides access to other databases (e.g., gene records), which are not of interest for this study. As a heuristic, we discarded sessions in which more than half the transactions did not involve MEDLINE; this removed an additional 2.72 m sessions. Ultimately, we ended up with a dataset containing 476k sessions, 7.65 m transactions; for clarity, we refer to this as the filtered dataset. Although the size of the dataset after applying these filters is a fraction of the original raw dataset, we argue that the processing steps allow us to focus on “interesting” user behavior, at least for the purposes of this study. Note that similar filtering is often applied to general Web query logs so that a researcher can focus on phenomena of interest. For example, previous analyses (Rose and Levinson 2004; Broder 2002) have found that, depending on the actual dataset, up to a quarter of all queries are navigational in nature, and up to a third of all queries are intended to locate a particular resource (e.g., finding a currency converter or downloading a video clip) rather than to gather information.
Building on Murray et al. (2006), the main idea explored in this work is to analyze user activities with n-gram language models; cf. (Chen and Cooper 2002). This is accomplished by first mapping classes of user actions into symbols drawn from a finite alphabet. Our encoding is shown in Table 1. Thus, sessions can be represented as sequences of symbols, and building computational models of users’ activities can be recast into a sequence modeling problem. For reference, Table 1 also shows the distribution of the 7.65 m user actions in the filtered dataset. The following are the beginnings of three sample sessions encoded according to our scheme:
Even disregarding details such as the actual query and the timestamp, it is possible to construct an understanding of user behavior in each of these cases. In the first example, the sequence of R’s indicate that the user examined four MEDLINE citations on the same result page. The long sequences of Q’s in the second session suggest that the user had difficulty formulating a good PubMed query. The sequence of N’s in the third example indicates that the user looked at several pages of results without selecting a MEDLINE citation to examine in detail, before finally giving up and issuing a different query.
3 Session segmentation
We first tackled the session segmentation problem. Note that there is no consistent definition of a “session” in the literature. For some researchers, sessions are semantic in nature. As an example, He and Göker (2000) proposed that “the start and end of a session are the points where the role behind a query changes”, which intimately ties the notion of a session to information-seeking tasks. For others, sessions are non-semantic constructs, defined purely in terms of temporal inactivity (Silverstein et al. 1999) or in terms of the granularity at which the log data is gathered (Jansen et al. 2000). We adopted the second approach, defining sessions in terms of browser cookies. Despite differences in terminology, there is nevertheless consensus that user activities can be segmented into “meaningful” units.
To reduce confusion, we use the term “episode” to refer to meaningful units of activities within sessions. An episode might, for example, correspond to a specific information-seeking task. Although the identification of these units is difficult without understanding the semantics of users’ queries, temporal information alone provides significant information (Catledge and Pitkow 1995; He and Goker 2000). That is, a system could segment a sequence of user actions into episodes based on periods of inactivity. This method is extremely fast (a pre-requisite for online processing) and sidesteps thorny issues of query semantics. In addition, methods based purely on temporal information do not raise privacy concerns, compared to methods that depend on the actual semantics of the user action (e.g., queries and clickthroughs).
What should this inactivity threshold be? Naturally, we face a tradeoff: the longer the duration, the more likely we are to create episodes that span multiple, possibly unrelated, information-seeking tasks. On the other hand, shorter durations may yield incomplete episode fragments. For example, if the user spends a long time reading a result page, and then returns to continue browsing additional search results, the system might infer multiple episodes if the threshold is too short.
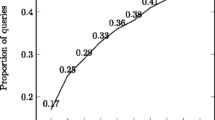
However, this tradeoff can be used to our advantage: in absence of “ground truth”, segmenting sessions into episodes based on different thresholds can serve as a probe into users’ behavior, by analyzing the characteristics of the resulting episodes. We did exactly this, segmenting sessions based on different inactivity thresholds, ranging from 5 to 60 min in 5 min increments. The line marked by diamonds in Fig. 1 denotes the total number of episodes that is generated at each threshold. Naturally, smaller thresholds generate more episodes. The line marked by filled squares represents the number of singleton episodes (i.e., episodes with only one transaction); the line marked by empty squares represents the number of singleton episodes consisting of a citation retrieval. The ratio between these two values is expressed as a percentage above the filled squares, i.e., the percentage of singleton episodes where the user retrieved a MEDLINE record. Note that from the citation view, users can access available full text (these actions are not recorded in our logs). We see that nearly 70% of all singleton episodes are retrievals.
Combined with the observation that about 60% of episodes end with a retrieval (regardless of threshold), we infer that singleton retrieval episodes are likely to be an extension of the previous episode—since the user is directly accessing MEDLINE content. By comparing these results with figures cited in Jansen and Spink (2004), we conclude that PubMed users spend a longer period of time examining content. This makes sense given the more complex tasks that PubMed users typically engage in—for example, physicians searching for clinical evidence in the context of patient care (De Groote and Dorsch 2003; Herskovic et al. 2007), biologists combing the literature for studies that link a particular gene to a disease (Hersh et al. 2005, 2007).
Figure 2 shows the distribution of episodes in terms of two different measures of length: number of transactions (top) and duration (bottom). For both graphs, we show the results of segmentation based on three representative thresholds: 5, 15, and 30 min. The plots for even longer thresholds are not substantially different from the plot for the 30-min threshold, and unfortunately showing any more lines would result in too much clutter. Duration is measured as the time difference between the first transaction of the episode and the last transaction (thus singleton episodes have zero duration). The bottom graph shows duration binned in increments of 5 min, e.g., increment 6 corresponds to all durations between 25 and 30 min long. In terms of number of interactions, the median episode length is 3 with an inactivity threshold of 5 and 10 min; 4 with a threshold of 15, 20, 25, 30 min; 5 with any longer thresholds. In terms of duration, the median episode length is less than 5 min for an inactivity threshold of 5, 10, 15, 20, 25, 30 min, rising to between 5 and 10 min for any longer thresholds. In both cases, the means are much larger than the medians since the distributions exhibit long tail characteristics.
Note that a meaningful comparison between PubMed data and data from general-purpose Web search engines (e.g., Google or Yahoo) is difficult, since most existing logs available to academic researchers contain only queries; see overview in Jansen and Spink (2006). In the case of PubMed, we have shown that episode boundaries cannot be accurately delineated without records of citation retrievals, since PubMed users may spend significant time examining MEDLINE citations.
These results seem to suggest that the choice of threshold is perhaps not as significant as one might think. With the exception of the 5-min threshold (which seems too short given the discussion above), the plots for episode length distribution don’t actually differ by much. In terms of transactions, longer thresholds are primarily reducing the number of singleton episodes, which are mostly citation retrievals. This has the effect of appending an additional ‘R’ symbol at the end of other sequences, and has a relatively minor effect for the experiments we describe in subsequent sections. Similarly, different thresholds have relatively minor impact on episode duration (disregarding the 5-min threshold).
To determine if there is any value in our simple notion of episodes and to better facilitate subsequent computational modeling, we prepared another dataset, which we refer to as the episode dataset. From the original set of filtered sessions we obtained a set of episodes by first segmenting the logs using a 30-min threshold. We then discarded the following:
-
all singleton episodes (since they cannot contribute to the user action prediction task we describe in Sect. 4);
-
all episodes that do not involve MEDLINE, i.e., consisting exclusively of the symbol ‘X’ (since our study focuses on searching MEDLINE);
-
all episodes that do not begin with a query (since they often represent direct access into MEDLINE, i.e., through another search engine, and we are primarily interested in studying search with PubMed).
From the 476k sessions in the filtered dataset, we ended up with 373k episodes containing 4.34 m transactions. For convenience, a summary of all datasets mentioned in this article is shown in Table 2.
4 Modeling user activities
One advantage of encoding user actions as sequences of symbols is the ability to use standard natural language processing techniques to build computational models of user activity. In this work, we experimented with n-gram language models. Language models define a probability distribution over string sequences:
In language processing, this typically means sequences of words; see Manning and Schütze (1999) for an overview. In our application, we are modeling sequences of symbols, each of which represent a user action. By the chain rule of probability theory:
Due to the extremely large number of parameters involved in estimating such a model, it is customary to make the Markov assumption, that the sequence histories only depend on prior local context. That is, an n-gram language model is equivalent to a (n − 1)-order Markov model. Thus, we can approximate P(w k |w k−11 ) as follows:
In this work, we used the SRI Language Modeling Toolkit (Stolcke 2002), a standard package commonly used by the computational linguistics and speech processing communities. Standard settings were used: Good-Turing discounting (Chen and Goodman 1996) and Katz backoff for smoothing (Katz 1987). For building the language models, we used both the filtered session data and the episode data, as summarized in Table 2. The datasets were divided in the following manner: for the filtered session dataset, a 400/76k training/test split; for the episode dataset, a 300/73k split. We varied the order of n-grams from 2-gram (bigrams) up to 8-gram.
In our first experiment, we built language models on training data and then evaluated these models on held-out test data. Cross entropy is frequently used as a metric:
where P m (t i ) denotes the probability assigned by language model m and where the test data T is composed of sequences (t 1, t 2,…,t n ) containing a total of N T symbols. The cross entropy is inversely related to the average probability a model assigns to sentences in the test data, and it is accepted that lower entropy models are preferable. Following common practices of the speech community, we report perplexity instead of cross entropy (H), where perplexity is 2H. One advantage is that the measure has an intuitive reading: a perplexity of k means that one is as surprised on average as one would have been guessing from k equiprobable choices at each symbol in the sequence.
Results for the perplexity experiments are shown in Fig. 3 for both filtered session data and episode data. We see that perplexity does decrease with higher-order models, although it appears that nothing is gained beyond 6-grams, and perplexity actually increases slightly due to the increasing sparsity of higher-order n-grams. Nevertheless, these results do suggest that there is regularity in sequences of user actions, and that this regularity can be modeled by taking into account history of previous actions.
In our second experiment, we applied the language models to a sequence prediction task. That is, after observing the user’s current history of actions, can the model “guess” what the user is going to do next? The prediction is performed as follows: given a sequence of actions, the system generates eight sequences, one corresponding to each possible next user action. These sequences are then scored by the language model; the user action that generates the most probable sequence is then predicted.
The experimental runs were structured in the following manner: a sequence of n user actions (either from a single session or episode, depending on the dataset) is divided into n − 1 trials. At each trial, the system’s task is to predict the next symbol, provided the history. Note that this experimental setup places higher-order n-gram models at a disadvantage, since for the first few predictions of any session or episode, information learned from higher-order n-grams cannot be exploited. However, this procedure does have the advantage that the number of trials is constant across all models.
Experiments were conducted using the language models built from the training data. From the session test data, we selected 10k sessions for testing prediction accuracy, which yielded 154,333 trials. From the episode data, we selected 20k episodes, which yielded 213,107 trials. We measured the accuracy of the predictions by comparing system output with the actual user actions—these results are shown in Fig. 4. The error bars denote the 99% confidence intervals, as computed by the Clopper-Pearson method for calculating exact binomial confidence intervals (Clopper and Pearson 1934). Due to the large number of trials, the confidence intervals are quite small, thus allowing us to discriminate small differences. In both graphs, the solid line indicates the baseline (frequency of most common class: 0.388 for episode, 0.397 for sessions).
Results from the prediction accuracy experiment are consistent with the perplexity evaluation. For both session and episode data, 5-gram models significantly outperform 4-gram and lower order models. The differences in prediction accuracy between 5-, 6-, 7-, and 8-gram models are not statistically significant for both session and episode data. To facilitate comparison, the scales of the bar graphs in Fig. 4 are the same. We can see that relative to the baseline, prediction accuracy is slightly higher on episode data than it is on session data: for example, 6-gram prediction accuracy for episodes is 0.503 or 30% above the baseline, while 6-gram prediction accuracy for sessions is 0.508 or 28% above the baseline. Recall that, by construction, the episode data contained more “interesting” sequences of user activities—these results suggest that there is some value in the episodes generated by our simple session segmentation technique.
5 Detecting activity collocates
By encoding users’ actions as string sequences, we can leverage well-known natural language processing techniques to identify interesting patterns. Computational linguists have devised a number of techniques for extracting collocations, or commonly-occurring patterns of words; see Pearce (2002) for a survey. Can such techniques be applied to help us better understand user behavior?
First, what would collocations in activity sequences derived from our logs mean? Collocations in natural language are co-occurring words that appear more frequently than one would expect by chance; often, their semantics are non-compositional (e.g., compound nouns, verb-particle constructions, etc.). In our context, collocations represent sequences of actions that are “surprising” and “meaningful”. These “activity collocates” might encapsulate fragments of information-seeking strategies or “idioms” that PubMed searchers have adopted, and analyzing such sequences could reveal interesting insights about user behavior. Depending on the absolute frequency of occurrence, these information-seeking strategies might be relatively common, or exclusively used by a small population.
To find activity collocates, we used Pointwise Mutual Information (Church and Hanks 1990) to score n-grams from our language models:
That is, PMI quantifies the probability of seeing a particular sequence relative to the probability that the individual symbols making up the sequence co-occurred randomly. We opted for PMI instead of more complex formulations such as the log likelihood ratio (Dunning 1993) since our alphabet is small and thus our modeling task is not plagued by problems associated with rare events; cf. (Moore 2004).
Table 3 shows the top five activity collocates in terms of PMI for 2-, 3-, 4-action sequences. Analysis was performed on the 400k training samples in the filtered session data, but the episode data gives rise to similar results. The table also shows the log probability of each n-gram to quantify the prevalence of that particular pattern. For reference, the most frequently-occurring patterns of activity are shown in Table 4. Note that in both cases, we discarded patterns that involved non-MEDLINE actions (the symbol ‘X’), since the probabilities of those actions are distorted by our data preparation process.
In the PubMed interface, the detailed view of a MEDLINE citation (which contains bibliographic information, and in most cases, abstract text) is accompanied by five links to related articles, as suggested by a probabilistic content-similarity algorithm (Lin and Wilbur 2007). Below the five titles is an option to see a longer list of related articles. Action ‘L’ represents a click on one of these suggested article titles; action ‘M’ represents a click on the option to see a longer list of related articles. Overall, ‘L’ and ‘M’ represent 3.7% and 0.7% of all transactions, respectively, yet most of the activity collocates involve these two actions. That is, these sequences occur far more frequently than one would expect by chance. Consecutive L’s indicate that the user was clicking on related article suggestions repeatedly (navigating from citation to citation); consecutive M’s indicate that the user was navigating from lists of related articles to other lists of related articles (essentially, using MEDLINE citations repeatedly as “queries” to retrieve lists of similar citations).
Our analysis suggests that browsing related articles (in its two forms) represents a deliberate information-seeking strategy, or more generally, a distinct way of using PubMed. Furthermore, once users begin browsing the collection in this manner, they are likely to continue—as shown by the long sequences of ‘L’ and ‘M’ actions in Table 3. Complementary evidence presented in Lin et al. (2008) supports an even stronger claim, that the feature is indeed useful and effective for information seeking. Through analysis of document networks connected via content-similarity links, Lin et al. showed that for typical information needs, relevant documents tend to cluster together. Therefore, a user can navigate from relevant document to relevant document via these links. Finally, these results are consistent with previous studies that demonstrate the effectiveness of content-similarity browsing in simulated environments (Wilbur and Coffee 1994; Smucker and Allan 2006; Lin and Smucker 2008). Although similar features are available in many Web search engines, we are not aware of any published evidence regarding their effectiveness.
The other activity collocates are comprised of N’s (next page) and P’s (other MEDLINE actions). The “N N” sequence suggests that users are often persistent in examining the retrieved set—that is, they browse through at least two pages of PubMed results (each of which contains 20 citations). A natural explanation is the recall-oriented nature of typical tasks that PubMed users engage in, e.g., physicians searching for clinical evidence or biologists searching for relevant literature. This behavior stands in contrast with general Web search, where users are much more cursory in their consumption of search results. For example, Jansen et al. (2000) observed that 58% of users don’t look past the first page of results (10 hits), and only around 10% of users view results past the fourth page.
As described in Table 1, the symbol ‘P’ serves as a catch-all category for otherwise uncategorized MEDLINE-related actions. These include actions in the “Preview/Index”, “History”, “Clipboard”, and “Details” tabs of the PubMed interface, as well as clicks on the “LinkOut” resource supplied by external providers. Examination of the logs reveals that sequences of P’s represent activities of advanced PubMed users. For example, the “Preview/Index” tab allows the user to see the number of hits that would be retrieved by a particular query—an important feature for Boolean retrieval since the result set size is often difficult to control. The “History” tab allows the user to revisit previously-issued queries. Overall, such actions are rare (only 3.8% of all page views in the filtered session dataset), but we observe consecutive P’s having high PMI. As with sequences of consecutive L’s and M’s, we believe that this represents a distinct mode of information seeking—for example, an advanced user encountering difficulty in choosing good search terms might switch to the “Preview/Index” tab as a tool for assisting in the query formulation process.
Focusing on Table 4, it is not surprising that the most frequent patterns of activity comprise mostly of ‘Q’ (query) and ‘R’ (retrieve) actions. Together, they account for nearly three quarters of total transactions. Sequences of these two actions represent perhaps the “core” of information-seeking behavior: issuing queries, examining results, and reformulating previous queries. We also note that activity collocates (i.e., those in Table 3) aren’t necessarily rare in terms of absolute frequency. For example, “N N” is the 5th most frequent 2-gram and “L L” is the 13th most frequent 2-gram.
Some frequent n-grams actually have surprisingly high PMI values. For example, we observe the prevalence of long query sequences. One possible interpretation is that at least some users experience difficulty formulating Boolean queries. Since almost all commercial Web search engines implement some sort of best-match algorithm, users have grown accustomed to using ranked retrieval systems. In contrast, the query formulation process in PubMed may feel quite foreign. In fact, based on analysis of a separate set of logs, we found that approximately a fifth of all PubMed queries return zero results. Related to these challenges is the difficulty associated with controlling the result set size, which is another characteristic of Boolean retrieval. For example, adding an additional term to a query that retrieves 1000 hits might yield 0 hits (this helps to explain the usefulness of the “Preview/Index” tab in the PubMed interface, for searchers who are aware of the functionality). These issues point to query formulation aids as features that could potentially benefit many PubMed users.
The sequences of consecutive repeated actions in both Tables 3 and 4 suggest an interesting behavior pattern: once a user commits to an action, he or she is likely to repeat the same action again. We can quantify this by computing P(L|L)/P(L); that is, how much more likely is action ‘L’ to follow another action ‘L’ (compared to chance)? The answer in this case is around 12 times more likely: ‘L’ follows another ‘L’ about 45% of the time. We computed similar values for longer sequences and for other actions, the results of which are shown in Fig. 5. Given the above discussion, the plots for action ‘L’ and action ‘N’ (next result page) are not unexpected: once users begin browsing related articles or pages in the result set, they are likely to continue doing so. However, we also observe the same effect for Query and Retrieve (albeit the effects are more minor): given a consecutive sequence of four ‘Q’ or ‘R’ actions, we are 2.07 and 1.86 more likely to see another ‘Q’ or ‘R’, respectively. This appears to support our characterization of PubMed users as “persistent”, which makes sense given the complex nature of their information needs.
6 Conclusion
In this work, we demonstrate that simple n-gram models can capture regularities in users’ activities, based only on a rough characterization of their actions. With these techniques, we are able to identify activity collocates and characterize interesting patterns of behavior. Overall, the models are able to correctly predict the user’s next action about half the time. We find this to be promising, considering the impoverished input—no temporal information (beyond pre-segmented sequences), no information about the semantics of the user’s actions (e.g., queries they issued, the contents of the pages they clicked on, etc.). Of course, models can be enriched by exactly such data to provide a more accurate picture of users’ behavior. However, the biggest barrier to this is not technological, but rather one related to policy: concerns over invasion of privacy and release of personally-sensitive information. For a discussion, see Murray and Teevan (2007). In this respect, log-mining techniques that are able to extract information from impoverished datasets are particularly valuable, since they alleviate these concerns. For academic researchers, it is more likely that such datasets can be shared.
While user modeling may be interesting in itself, our ultimate goal is to exploit such models to improve the search experience. We believe that user models can be used in three major ways:
-
Predictively. In anticipation of what the user is likely to do next, the system can provide customized assistance. One might consider this as a generalization of query suggestion techniques that have been proposed (Anick 2003; Cui et al. 2003). That is, in addition to suggesting related search terms, the system might offer helpful actions. For example, activity collocates might be generalized into a library of “search strategies” and presented as search aids. For example, the system may suggest something like, “similar users have found related article links to be helpful—perhaps you might want to consider…”
-
Retrospectively. Computational models can help systems automatically cluster and classify user behavior. Results in Sect. 4 offer some possibilities for how this might be accomplished. The biggest application is user profiling and demographic modeling, which is highly relevant to many tasks ranging from personalized search (Eirinaki and Vazirgiannis 2003; Shen et al. 2005) to targeted advertisement.
-
Prescriptively. That is, results of log-based studies could be used as a basis for educating users on effective search strategies. This is not an unrealistic scenario in the context of PubMed: due to the nature of its users and their work, PubMed searchers are often willing to learn effective search techniques and advanced features. Footnote 2
We believe that this work represents an enabler for such advances. Transaction logs help us better understand how PubMed users search for information and provide a valuable resource for building computational models of user activities. We demonstrated how user actions can be encoded as string sequences and captured with n-gram language models. The application of natural language processing techniques to tackle this information retrieval problem provides an example of how the two fields might productively collaborate.
Notes
Empirical evidence for this claim is demonstrated by the numerous tutorials and mini-courses offered on PubMed, as any casual Web search will reveal.
References
Agichtein, E., Brill, E., & Dumais, S. (2006). Improving Web search ranking by incorporating user behavior information. In Proceedings of the 29th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR 2006) (pp. 19–26). Seattle, WA.
Anick, P. (2003). Using terminological feedback for Web search refinement—A log-based study. In Proceedings of the 26th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR 2003) (pp. 88–95). Toronto, Canada.
Beitzel, S. M., Jensen, E. C., Chowdhury, A., Grossman, D., & Frieder, O. (2004). Hourly analysis of a very large topically categorized Web query log. In Proceedings of the 27th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR 2004) (pp. 321–328). Sheffield, UK.
Broder, A. (2002). A taxonomy of Web search. SIGIR Forum, 36(2), 3–10.
Cahan, M. A. (1989). GRATEFUL MED: A tool for studying searching behavior. Medical Reference Services Quarterly, 8(4), 61–79.
Catledge, L. D., & Pitkow, J. E. (1995). Characterizing browsing strategies in the World-Wide Web. Computer Networks and ISDN Systems, 27(6),1065–1073.
Chen, H.-M., & Cooper, M. D. (2002). Stochastic modeling of usage patterns in a Web-based information system. Journal of the American Society for Information Science and Technology, 53(7), 536–548.
Chen, S. F., & Goodman, J. (1996). An empirical study of smoothing techniques for language modeling. In Proceedings of the 34th Annual Meeting of the Association for Computational Linguistics (ACL 1996) (pp. 310–318). Santa Cruz, CA.
Church, K. W., & Hanks, P. (1990). Word association norms, mutual information, and lexicography. Computational Linguistics, 16(1), 22–29.
Clopper, C. J., & Pearson, E. S. (1934). The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika, 26.404–413.
Cui, H., Wen, J.-R., Nie, J.-Y., & Ma, W.-Y. (2003). Query expansion by mining user logs. IEEE Transactions on Knowledge and Data Engineering, 15(4), 829–839.
De Groote, S. L., & Dorsch, J. L. (2003). Measuring use patterns of online journals and databases. Journal of the Medical Library Association, 91(2), 231–240.
Dunning, T. (1993). Accurate methods for the statistics of surprise and coincidence. Computational Linguistics, 19(1), 61–74.
Eirinaki, M., & Vazirgiannis, M. (2003). Web mining for Web personalization. ACM Transactions on Internet Technology, 3(1), 1–27.
Haynes, R. B., Wilczynski, N., McKibbon, K. A., Walker, C. J., & Sinclair, J. C. (1994). Developing optimal search strategies for detecting clinically sound studies in MEDLINE. Journal of the American Medical Informatics Association, 1(6), 447–458.
He, D., & Göker, A. (2000). Detecting session boundaries from Web user logs. In Proceedings of the BCS-IRSG 22nd Annual Colloquium on Information Retrieval Research (pp. 57–66). Cambridge, UK.
Hersh, W. R., Cohen, A., Ruslen, L., & Roberts, P. (2007). TREC 2007 Genomics Track overview. In Proceedings of the Sixteenth Text REtrieval Conference (TREC 2007). Gaithersburg, MD.
Hersh, W. R., Cohen, A., Yang, J., Bhupatiraju, R., Roberts, P., & Hearst, M. (2005). TREC 2005 Genomics Track overview. In Proceedings of the Fourteenth Text REtrieval Conference (TREC 2005), Gaithersburg, MD.
Herskovic, J. R., Tanaka, L. Y., Hersh, W. R., & Bernstam, E. V. (2007). A day in the life of PubMed: Analysis of a typical day’s query log. Journal of the American Medical Informatics Association, 14(2), 212–220.
Horowitz, G. L., Jackson, J. D., & Bleich, H. L. (1983). PaperChase. Self-service bibliographic retrieval. JAMA, 250(18), 2494–2499.
Jansen, B. J., & Spink, A. (2004). An analysis of documents viewing patterns of Web search engine users. In A. Scime (Ed.), Web mining: Applications and techniques (pp. 339–354). Hershey, PA: IGI Publishing.
Jansen, B. J., & Spink, A. (2006). How are we searching the World Wide Web? A comparison of nine search engine transaction logs. Information Processing and Management, 42(1), 248–263.
Jansen, B. J., Spink, A., & Saracevic, T. (2000). Real life, real users, and real needs: A study and analysis of user queries on the Web. Information Processing and Management, 36(2), 207–227.
Joachims, T., Granka, L., Pan, B., Hembrooke, H., Radlinski, F., & Gay, G. (2007). Evaluating the accuracy of implicit feedback from clicks and query reformulations in Web search. ACM Transactions on Information Systems, 25(2), 1–27.
Katz, S. M. (1987). Estimation of probabilities from sparse data for the language model component of a speech recognizer. IEEE Transactions on Acoustics, Speech, and Signal Processing, 35(3), 400–401.
King, N. S. (1991). Search characteristics and the effects of experience on end users of PaperChase. College and Research Libraries, 52(4), 360–374.
Lin, J., DiCuccio, M., Grigoryan, V., & Wilbur, W. J. (2008). Navigating information spaces: A case study of related article search in PubMed. Information Processing & Management, 44(5), 1771–1783.
Lin, J., & Smucker, M. D. (2008). How do users find things with PubMed? Towards automatic utility evaluation with user simulations. In Proceedings of the 31st Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR 2008) (pp. 19–26). Singapore.
Lin, J., & Wilbur, W. J. (2007). PubMed related articles: A probabilistic topic-based model for content similarity. BMC Bioinformatics, 8, 423.
Manning, C. D., & Schütze, H. (1999). Foundations of statistical natural language processing. Cambridge, MA: MIT Press.
Moore, R. C. (2004). On log-likelihood-ratios and the significance of rare events. In Proceedings of the 2002 Conference on Empirical Methods in Natural Language Processing (EMNLP 2002) (pp. 333–340). Barcelona, Spain.
Murray, G. C., Lin, J., & Chowdhury, A. (2006). Action modeling: Using language models to predict query behavior. In Proceedings of the 29th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR 2006) (pp. 681–682). Seattle, WA.
Murray, G. C., & Teevan, J. (2007). Query log analysis: Social and technological challenges. SIGIR Forum, 41(2), 112–120.
Pearce, D. (2002). A comparative evaluation of collocation extraction techniques. In Proceedings of the Third International Conference on Language Resources and Evaluation (LREC 2002) (pp. 1530–1536). Canary Islands, Spain.
Rose, D. E., & Levinson, D. (2004). Understanding user goals in Web search. In Proceedings of the Thirteenth International World Wide Web Conference (WWW 2004) (pp. 13–19). New York, NY.
Shen, X., Tan, B., & ChengXiang, Z. (2005). Implicit user modeling for personalized search. In Proceedings of the 2005 ACM CIKM International Conference on Information and Knowledge Management (pp. 824–831). Bremen, Germany.
Silverstein, C., Marais, H., Henzinger, M., & Moricz, M. (1999). Analysis of a very large Web search engine query log. SIGIR Forum, 33(1), 6–12.
Smucker, M. D., & Allan, J. (2006). Find-similar: Similarity browsing as a search tool. In Proceedings of the 29th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR 2006) (pp. 461–468). Seattle, WA.
Stolcke, A. (2002). SRILM—An extensible language modeling toolkit. In Proceedings of the 7th International Conference on Spoken Language Processing (ICSLP 2002) (pp. 901–904). Denver, CO.
Wilbur, W. J., & Coffee, L. (1994). The effectiveness of document neighboring in search enhancement. Information Processing and Management, 30(2), 253–266.
Acknowledgments
We’d like to thank three anonymous reviewers for their helpful comments. This work was supported by the Intramural Research Program of the NIH, National Library of Medicine. The first author would also like to thank Esther and Kiri for their kind support.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lin, J., Wilbur, W.J. Modeling actions of PubMed users with n-gram language models. Inf Retrieval 12, 487–503 (2009). https://doi.org/10.1007/s10791-008-9067-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10791-008-9067-7