Abstract
We report results of a study that made reciprocal comparisons of environmental DNA (eDNA) assays for two major invasive crayfishes between their disparate invasive ranges in North America. Specifically, we tested for range expansions of the signal crayfish Pacifastacus leniusculus (Dana, 1852) into the Laurentian Great Lakes region known to be invaded by the rusty crayfish Orconectes rusticus (Girard, 1852), as well as for the invasion of O. rusticus into large lakes of California and Nevada, US known to be invaded by P. leniusculus. We compared eDNA detections to historic localities for O. rusticus within the Great Lakes, and to recent sampling for presence/absence and relative abundance of P. leniusculus in California and Nevada via overnight sets of baited traps. We successfully detected O. rusticus eDNA at six sites from the Great Lakes and P. leniusculus from six of seven lakes where it was known to occur in California and Nevada, but did not detect any range expansions by either species across the North American continent. eDNA appears suitable to detect benthic arthropods from exceptionally large lakes, and will likely be useful in applications for monitoring of new biological invasions into these and other freshwater and marine habitats.
Similar content being viewed by others
Introduction
Biological invasions profoundly affect aquatic ecosystems and their constituent organisms globally (Gallardo et al., 2016), and consequently demand ongoing development of tools for more effective prevention and control (Lodge et al., 2016). This includes surveillance approaches that are sensitive enough to detect new invasions early when eradication or containment efforts are likely to be most successful and cost-effective (Simberloff, 2003; Vander Zanden et al., 2010). Among the most promising advances in invasive species surveillance to emerge recently has been environmental DNA (eDNA), or DNA of macrobiota collected and identified from environmental samples (Ficetola et al., 2008; Lodge et al., 2012b). Research to date has consistently found eDNA to be highly sensitive to detection of potentially harmful species at the low population abundances associated with early stages of invasion (e.g., Egan et al., 2015; Smart et al., 2015; Dougherty et al., 2016; Matsuhashi et al., 2016). However, a number of questions related to the application and interpretation of eDNA methods persist and require ongoing research attention (Roussel et al., 2015; Barnes & Turner, 2016).
Among these concerns is the performance or suitability of eDNA for applications to more diverse taxa and habitats beyond the fish or amphibians and small ponds or mesocosms where this tool was initially pioneered (e.g., Ficetola et al., 2008; Goldberg et al., 2011; Barnes et al., 2014). As examples, the performance of eDNA for benthic arthropods like freshwater crayfish has been equivocal between initial studies (e.g., Tréguier et al., 2014; Figiel & Bohn, 2015; Dougherty et al., 2016), and tests of eDNA in the largest of freshwater habitats like the Laurentian Great Lakes of the United States (US) and Canada (hereafter Great Lakes) have been relatively rare (but see Tucker et al., 2016). Given that organisms like crayfish have invaded and negatively affected large freshwater habitats including the Great Lakes (Peters et al., 2014), we ask: is it feasible to provide early detection of invasive benthic arthropods in massive freshwater lakes using eDNA? We addressed this question by developing and applying eDNA assays for two major invasive crayfishes in large lakes of their reciprocal non-native ranges in North America. Our study allowed us to both test the feasibility of eDNA for detecting benthic arthropods in situ in large freshwater ecosystems while simultaneously screening for major range expansions of two problem species into new, potentially suitable regions.
The rusty crayfish Orconectes rusticus (Girard, 1852) and the signal crayfish Pacifastacus leniusculus (Dana, 1852) are two of the most widespread and impactful invasive crayfishes on the planet (Lodge et al., 2012a); their ecological effects include dramatically reducing populations of aquatic plants, other macroinvertebrates, native crayfishes, and some imperiled vertebrate species (Twardochleb et al., 2013). O. rusticus is native to the Ohio River drainage in the US, but was introduced northward through pathways including live bait use by anglers into the Great Lakes, as well as inland lakes and rivers of US states and Canadian provinces like Wisconsin and Ontario (Capelli & Magnuson, 1983; Edwards et al., 2009; Peters et al., 2014). More recently, O. rusticus has established its first populations in western North America (e.g., Olden et al., 2009), and species distribution modeling suggests that more of the western US is likely vulnerable to further invasion by O. rusticus (Morehouse & Tobler, 2013). Conversely, P. leniusculus is native to the Columbia River and adjacent Pacific drainages of western North America, but was introduced over the past century southward to the US states of California and Nevada, and then subsequently to Europe and Japan (Lodge et al., 2012a; Larson & Williams, 2015; Usio et al., 2016). Species distribution models predict that much of eastern North America—including the Great Lakes—is likely suitable for P. leniusculus to establish populations, although this invasive crayfish has not been previously observed from this half of the continent (Capinha et al., 2011; Larson & Olden, 2012).
We sought to evaluate whether eDNA could detect these two invasive crayfishes from among the largest freshwater habitats of their non-native ranges, using reciprocal surveillance efforts between the Great Lakes (O. rusticus established, P. leniusculus absent but at risk for invasion) and large lakes of California and Nevada (P. leniusculus established, O. rusticus absent but at risk for invasion). We used a quantitative PCR (qPCR) eDNA assay that was recently found effective at detecting O. rusticus in smaller inland lakes (Dougherty et al., 2016), and developed a new qPCR eDNA assay for P. leniusculus using a recently published, comprehensive molecular dataset on the phylogenetics of the Pacifastacus genus (Larson et al., 2016). We collected water samples for eDNA from locations across all five of the Great Lakes and several associated waters, including Lake Champlain (133,100 ha) and the St. Lawrence Seaway, as well as from the two largest natural lakes in California (Lake Tahoe at 49,000 ha, Clear Lake at 18,000 ha) and a number of nearby, smaller lakes and reservoirs. We also used citizen scientists in affiliation with public zoo and aquarium summer science programs to collect some water samples from the Great Lakes region. Finally, we compared eDNA detections for O. rusticus to recent distributional records for this species from a comprehensive review of crayfish across the Great Lakes (Peters et al., 2014), and compared eDNA detections for P. leniusculus to results of baited trapping for this species in a subset of the California and Nevada lakes. Cumulatively, our multi-region study tested whether eDNA can successfully detect invasive benthic arthropods from large lake habitats while screening for new potential crayfish invasions across the North American continent.
Methods
Our study included (a) design and testing of primers to amplify P. leniusculus DNA, (b) collecting water samples from the field in both the Great Lakes and California and Nevada study regions, (c) running single-species qPCR assays for both crayfish species on samples from both study regions, and finally (d) comparing eDNA detections to either known distributions (O. rusticus) or previous sampling for relative abundance (P. leniusculus) to evaluate the performance of eDNA for these crayfish.
Primer design and testing
We used a primer pair designed for the O. rusticus cytochrome c oxidase subunit I (COI) mitochondrial DNA gene previously published by Dougherty et al. (2016) that was tested for specificity against tissue samples from all other known Great Lakes region sympatric crayfishes (11 species), and that was also found effective at detecting this invasive crayfish from inland lakes of Wisconsin and Michigan, US. We refer readers to Dougherty et al. (2016) for more details on primer design, testing, and previous performance, but report here that the primer pair used was Orusticus_COI_5F (5′-CAGGGGCGTCAGTAGATTTAGGTAT-3′) and Orusticus_COI_5R (5′-CATTCGATCTATAGTCATTCCCGTAG-3′), which produces a 128 base pair (bp) amplicon.
We designed and tested a new primer pair to amplify a subregion of the COI gene for P. leniusculus. We designed primers using Primer3 (Untergasser et al., 2012) and visual searches for nucleotide variants between in-group P. leniusculus sequences and out-group sequences of other crayfishes of the genus Pacifastacus and family Astacidae. The best primer pair we could identify, PacifastacusE_COI_F2 (5′-GGRGGATTTGGTAATTGGTTAATTC-3′) and PacifastacusE_COI_R2b (5′-CAATAGCCGCTGCTAGAGGA-3′), produced a 184 bp amplicon. Specificity of the above primer pair was evaluated in the lab for successful amplification of tissue-derived P. leniusculus genomic DNA, as well as reduced amplification with tissue-derived genomic DNA for 2 other crayfish species of the genus Pacifastacus, 4 of the European crayfish species of the family Astacidae, and 10 North American crayfish species of the family Cambaridae known to occur in the Great Lakes region, including O. rusticus. We evaluated performance of our primer pair using qPCR settings consistent with those reported in processing of our field samples (below). We required both a qPCR amplification curve and proper melting curve relative to that of the targeted in-group taxa to consider a sample positive, and also evaluated the timing of the quantification cycle (Cq) when fluorescence from amplification exceeds background fluorescence to further evaluate the strength of a non-target tissue-derived amplification as compared to amplification from target tissue-derived samples. Primer testing results confirmed our assay as specific enough to discriminate P. leniusculus DNA from non-target species, although we recommend post-qPCR Sanger sequencing confirmation. Additional details on P. leniusculus primer design and testing, particularly related to the complex taxonomy and phylogeny of the Pacifastacus genus (Larson et al., 2012, 2016), are available in Electronic Supplementary Material.
Field sample collection
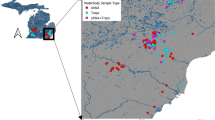
We collected eDNA field samples from 14 sites within the Great Lakes region between 8 June 2015 and 22 October 2015, although a majority of samples were taken in the months of July and August (Electronic Supplementary Table S3). We collected eDNA samples from all five of the Great Lakes themselves, as well as from sites further downstream in the watershed in Lake Champlain and the St. Lawrence Seaway (Fig. 1). Specific site locations with sample dates and geographic coordinates and their abbreviations used throughout the manuscript (e.g., Fig. 1) are provided in Electronic Supplementary Table S3. These sites included a mix of shorelines of both densely populated urban centers (e.g., Chicago, Toronto) where releases of invasive crayfishes new to the Great Lakes by humans might be likely (Capinha et al., 2013), as well as more remote locations where the risk of crayfish introductions might be lower (e.g., Apostle Islands National Lakeshore of Lake Superior, Beaver Island of Lake Michigan). Further, at 9 of these 14 sites, eDNA samples were taken by citizen scientists, either lead author-supervised or unsupervised, who were participating in public zoo or aquarium summer programs, which included 24 organization staff or employees and 70 student or teacher volunteers. Participating organizations and citizen scientists are listed in Electronic Supplementary Table S3. In two cases where unsupervised citizen scientist groups took eDNA water samples, prior training on the field sampling protocol had been provided to organization staff by the authors.
Location of eDNA sample sites for the rusty crayfish Orconectes rusticus and signal crayfish Pacifastacus leniusculus in both the Great Lakes and California and Nevada regions of North America, with sample site abbreviations (Electronic Supplementary Table S3), whether or not O. rusticus or P. leniusculus eDNA was detected at a site, and historic localities for O. rusticus within the Great Lakes from Peters et al. (2014)
We collected eDNA field samples from 11 lakes or reservoirs in California and Nevada between 26 and 31 August 2015 (Fig. 1). These sites included the two largest natural lakes in California (Lake Tahoe and Clear Lake), as well as a series of smaller lakes and reservoirs (Electronic Supplementary Table S3). We sampled from seven locations around the perimeter of Lake Tahoe and from both the southern and western shoreline of Clear Lake, whereas most other study sites—regardless of region—had all replicated eDNA samples (below) taken from close proximity (within ~100 m) to a single location on their shoreline. Lake Tahoe in particular was chosen because of its history as a source for invasive P. leniusculus populations shipped to Europe (Larson & Williams, 2015), capacity to serve as a western North American surrogate for the Great Lakes in a cross-continental comparison, and finally for the availability of recent monitoring of P. leniusculus relative abundance throughout this lake (and some of its neighbors) for comparison to our eDNA results. Further details on recent P. leniusculus monitoring as related to eDNA results are addressed later in the methods.
In both the Great Lakes and California and Nevada sampling regions, we took 10 replicated eDNA water samples at each sample site, with the exception of instead taking 5 replicated eDNA water samples at the spatially dispersed sites in Lake Tahoe (Electronic Supplementary Table S3). Surface water samples of 250 ml volume were taken in bottles that had previously been decontaminated by minimum 10-min soaks in 10% bleach solution prior to a rinse in deionized water (Goldberg et al., 2016). These water samples were then immediately filtered through funnels containing 1.2 μm cellulose nitrate filters using a hand vacuum pump (Actron CP7830; Bosch Automotive Service Solutions, Warren, Michigan, US) connected to a side-arm flask. Filters were then promptly placed in 2-ml microcentrifuge tubes (USA Scientific, Ocala, Florida, US) and completely submerged in 700 μl of Longmire’s buffer (Longmire et al., 1997). To test for contamination of our field eDNA filtering approach, we processed as above controls or blanks of 250 ml sample volumes of store-bought, bottled water for every five field samples (two controls per sites with 10 eDNA water samples). Nitrile gloves were worn and changed between individual samples for both water collection and filtering steps. Filtered field samples and controls were shipped to the University of Notre Dame, South Bend, Indiana, US where they were stored in a refrigerator prior to eDNA extraction from filters and buffer in autumn of 2015 and winter of 2016. Storage in Longmire’s buffer has been found to reliably preserve eDNA concentrations from filtered field samples at room temperature or cooler for at least 150 days (Renshaw et al., 2015; Wegleitner et al., 2015).
Laboratory eDNA sample processing
All eDNA extractions followed a modified chloroform–isoamyl alcohol (hereafter “CI”) DNA extraction and isopropanol precipitation protocol outlined in Renshaw et al. (2015): (1) 2-ml microcentrifuge tubes were incubated in a 65°C water bath for a minimum of 10 min, (2) 700 µl of CI (24:1, Amresco) was added to each tube and samples were vortexed for 5 s, (3) tubes were centrifuged at 15,000×g for 5 min and 500 µl of the aqueous layer was transferred to a fresh set of 1.5 ml microcentrifuge tubes, (4) 500 µl of ice cold isopropyl alcohol and 250 µl of 5 M NaCl were added to the 500 µl removed from the aqueous layer and tubes were precipitated at −20°C overnight, (5) the precipitate was pelleted by centrifugation at 15,000×g at room temperature for 10 min and the liquid was decanted, (6) 150 µl of room temperature 70% ethanol was added to each tube to wash pellets, (7) tubes were centrifuged at 15,000×g at room temperature for 5 min and the liquid was decanted, (8) 150 µl of room temperature 70% ethanol was added to each tube to wash pellets a second time, (9) tubes were centrifuged at 15,000×g at room temperature for 5 min and the liquid was decanted, (10) pellets were dried in a vacufuge at 45°C for 15 min, followed by air drying until no visible liquid remained, and finally, (11) pellets were rehydrated with 100 µl of 1× TE buffer, low EDTA (USB).
Three qPCR replicates were run for each eDNA extract in the following 20 µl reactions: 4.85 µl of PCR-grade water, 4 µl of 5× colorless GoTaq® flexi buffer (Promega), 0.4 µl of 10 mM dNTPs, 1.6 µl of 25 mM MgCl2, 1 µl of each 10 µM primer (forward and reverse), 0.15 µl of GoTaq® flexi DNA polymerase (Promega), 1 µl of EvaGreen (20× in water; Biotium), 2 µl of 4 µg/µl bovine serum albumin (Amresco), and 4 µl of eDNA extract. Mastercycler® ep realplex (Eppendorf) cycling conditions were as follows: an initial denaturation at 95°C for 3 min; 45 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 1 min, followed by a melting curve analysis that transitioned from 60 to 95°C over a span of 20 min. For quantification of eDNA samples, we used a 500 bp gBlock gene fragment based on GenBank accession AY701249 for O. rusticus, and a 487 bp gBlock gene fragment based on GenBank accessions EU921148, JF437995, and JF437997 for P. leniusculus. All gBlock gene fragments were synthesized by IDT. Copy numbers for gBlock fragments were estimated by multiplying Avogadro’s number by the number of moles. A serial dilution of the gBlock fragment provided a range in copy numbers for the quantification of eDNA unknowns (Gunawardana et al., 2014; Renshaw et al., 2015; Svec et al., 2015). We note as well that Great Lakes samples were run using both primers for O. rusticus known to occur there and for P. leniusculus being screened for new invasions, as well as the inverse (California and Nevada samples run twice using primers for both species).
Beyond our use of field controls to test for contamination, we also checked the eDNA extraction reagents and technique for contamination by the inclusion of a single extraction blank (one per each set of extracted eDNA samples) that involved just the reagents. On each qPCR assay, we checked the assay reagents and technique for contamination with two wells that included the same Mastermix as the rest of the plate with sterile water in place of the eDNA extract. The serial dilution of standards on each plate served as a qPCR-positive control. Finally, where amplification curves with the correct Cq and melting curves consistent with the target species (whether O. rusticus or P. leniusculus) were observed, a single qPCR replicate of these presumed positive eDNA detections was further confirmed through unidirectional Sanger sequencing with the reverse primer.
Comparison of eDNA to crayfish distributions or abundance
Unlike some previous studies including Dougherty et al. (2016) or Doi et al. (2016), we did not couple our eDNA field sampling with simultaneous conventional surveys for our target taxa. However, in both study regions, we had access to resources that could be used to relate eDNA results to either best available estimates of species distributions or recent measures of absence and relative abundance. First, Peters et al. (2014) recently summarized all known crayfish presence localities throughout the Great Lakes, using a combination of published academic and unpublished grey literature and government agency monitoring records (Fig. 1). While Peters et al. (2014) does not include reliable, widespread absence records for crayfish species such as O. rusticus or estimates of their population sizes or relative abundances in the Great Lakes, we still sought to relate our eDNA detections to distance to nearest observed O. rusticus presence locality. We did so with the expectation that some absences we observed for O. rusticus by eDNA might be the product of sampling sites that have not yet experienced O. rusticus invasion, and Peters et al. (2014) is the best available resource for this comparison. Similarly, we wanted to evaluate whether detections of O. rusticus eDNA tended to occur at sites either known to have been invaded by this crayfish or in close proximity to O. rusticus populations that may have spread to our sampling location recently. We estimated Euclidean distance in km from our eDNA sampling locations to the nearest O. rusticus records reported in Peters et al. (2014; Fig. 1), and tested for significant differences in distances between sites where O. rusticus eDNA was and was not detected. This analysis focused only on the 12 sample sites within the Great Lakes themselves and omitted 2 eDNA sample sites downstream in Lake Champlain and the St. Lawrence Seaway owing to the more limited geographic coverage of Peters et al. (2014).
In the California and Nevada region, 9 of our 11 study lakes had been recently sampled for P. leniusculus relative abundance as part of routine lake monitoring by the authors (Electronic Supplementary Table S4). This included Lake Tahoe and eight lakes in its immediate vicinity, but did not include the more remote Clear Lake or Camp Far West Reservoir (Fig. 1). Monitoring for invasive P. leniusculus in these lakes involves overnight sets of wire mesh crayfish traps (0.42 m long × 0.21 m diameter with two 60 mm diameter openings) over a depth gradient baited with dry dog food. Baited trapping results are reported as catch-per-unit effort (CPUE), the average number of crayfish collected per trap from the sampling event. Performance of baited trapping for detecting presence and reflecting relative abundance of crayfish in lakes is reviewed in Larson & Olden (2016). Monitoring of P. leniusculus populations within Lake Tahoe by baited trapping occurred at seven locations dispersed around the lake perimeter to which sample sites for eDNA were deliberately matched (Fig. 1). Seven of nine lakes previously sampled for crayfish by baited trapping were found to contain P. leniusculus of varying relative abundance, whereas two lakes have never had crayfish populations detected. Similarly, within Lake Tahoe, a gradient of P. leniusculus relative abundance from scarce to extremely abundant was known a priori, in response to poor (sandy) versus good (rocky) benthic habitats for this crayfish (Larson & Olden, 2013). We used linear regression to relate average eDNA copy number from qPCR to recent average P. leniusculus CPUE for each of the nine lakes individually, as well as for the seven locations within Lake Tahoe. This analysis allowed us to evaluate whether eDNA results agreed with relative abundance estimated from a more conventional sampling method over gradients of P. leniusculus prevalence both between lakes, as well as within one large lake. Additional details on baited trapping data used in this analysis are reported in Electronic Supplementary Table S4.
Results
We detected O. rusticus eDNA at 6 of 14 locations in the Great Lakes, and did not detect P. leniusculus eDNA anywhere in this region (Figs. 1, 2). eDNA concentrations for O. rusticus were highest in the vicinity of Milwaukee and Chicago, where 10 out of 10 water samples contained O. rusticus eDNA, and were intermediate at Grand Traverse Bay of Lake Michigan and Toronto. Conversely, O. rusticus eDNA was most scarce in Cleveland and Detroit, where 2 and 1 of 10 water samples, respectively, were found to contain low copy numbers of O. rusticus eDNA. eDNA of O. rusticus was not detected from lower in the watershed in Lake Champlain and the St. Lawrence Seaway (Figs. 1, 2).
Average eDNA copies per 4 μl + 1 on a log10 axis for both the Great Lakes (a) and California and Nevada (b) regions for the rusty crayfish Orconectes rusticus and signal crayfish Pacifastacus leniusculus. No O. rusticus eDNA was detected from California and Nevada lakes, and reciprocally no P. leniusculus eDNA was detected from the Great Lakes region. Sample site abbreviations are given in Electronic Supplementary Table S3 (see also Fig. 1). The number of positive detections per total number of eDNA water samples at each site is given below the x-axis of each plot
Pacifastacus leniusculus eDNA was detected at 6 of 11 lakes in California and Nevada, with high detections and copy numbers at most sites, excepting Prosser Reservoir where 2 of 10 water samples contained low eDNA copy numbers for this species (Figs. 1, 2). We did not detect O. rusticus eDNA from any California and Nevada lakes. We failed to detect P. leniusculus eDNA from one lake (Stampede Reservoir) known to previously support populations of this crayfish, but non-detections of P. leniusculus eDNA agreed with past sampling at two lakes believed to have no crayfish (Electronic Supplementary Table S4). We did not detect P. leniusculus eDNA from Clear Lake and Camp Far West Reservoir, which have not been sampled for crayfish by conventional (i.e., baited trapping) methods.
Finally, across qPCR runs for both species and regions, we found no evidence of contamination in either our field or laboratory controls. Across all sample runs, we observed average qPCR efficiencies of 99% (range 95–100%) and average R 2 values of 0.99 (range 0.92–1.00). All positive detections for O. rusticus and P. leniusculus were confirmed by Sanger sequencing and resulting match to available COI sequences for these species.
Proximity to Great Lakes O. rusticus localities
All O. rusticus eDNA detections in the Great Lakes were within 33.8 km of previously observed localities for this crayfish, whereas the majority of non-detections for O. rusticus were at sites where O. rusticus localities were more remote (52.1–100.0 km; Fig. 3). However, two localities (Duluth, Tawas Point on Lake Huron) where we did not detect O. rusticus eDNA were within 9.4 km or closer of recent historic collections of O. rusticus physical specimens (years 1999–2002). Accordingly, we did not observe any significant difference in distance to historically observed O. rusticus localities between sampling sites where we did and did not detect O. rusticus eDNA (Mann–Whitney rank sum U = 10.000, P = 0.240). We used a Mann–Whitney rank sum test for this comparison because of unequal variances between our two categories (Brown–Forsythe test P < 0.05).
eDNA copy number and P. leniusculus relative abundance
We found positive but weak relationships between eDNA copy number for P. leniusculus and relative abundance as CPUE for this crayfish estimated from recent baited trapping both in a nine lake subset of our California and Nevada study region, as well as for the seven locations within Lake Tahoe itself (Fig. 4). Again, we did not detect P. leniusculus eDNA from two lakes believed to have no crayfish populations, but also we did not detect P. leniusculus eDNA from one lake with notably high P. leniusculus CPUE values from recent sampling by baited trapping. Within Lake Tahoe, eDNA copy numbers were lowest at the one site within the lake known to have the lowest CPUE for this crayfish, but there was little relationship between eDNA copy number and crayfish relative abundance at the other six sites. In both cases, low R 2 values (0.151–0.155) evidence little relationship between P. leniusculus CPUE and eDNA copy number.
Relationships between median eDNA copies per sample + 1 (from Fig. 2) for the signal crayfish Pacifastacus leniusculus relative to the most recent estimates of relative abundance for this crayfish as catch-per-unit effort (CPUE) from overnight baited trapping at the same lakes (a) or locations within Lake Tahoe (b). Sample site abbreviations are given in Electronic Supplementary Table S3 (see also Fig. 1) and CPUE data are available in Electronic Supplementary Table S4
Discussion
Our study demonstrated that eDNA can detect benthic arthropods such as crayfishes in exceptionally large freshwater habitats, and also tested whether two major invasive crayfishes had expanded their ranges to climatically suitable regions on the North American continent. We were relieved to not find O. rusticus eDNA from California or Nevada, where it has never been directly observed, and also by our reciprocal failure to find P. leniusculus eDNA from the Great Lakes region. Further, within the known invasive range of each crayfish, eDNA generally performed well in comparison to either historic localities for O. rusticus within the Great Lakes or to lakes with and without known populations for P. leniusculus in California and Nevada. However, applications in each range had some apparent errors of omission or false negatives where other data sources indicated that the target crayfish species was present but it was not detected with eDNA. Finally, consistent with some past studies like Dougherty et al. (2016), we found weak relationships between eDNA copy number for the crayfish P. leniusculus relative to more conventional estimates of relative abundance for this species. More work is needed to determine the conditions over which eDNA can faithfully reflect patterns of relative abundance for benthic arthropods such as crayfish and other taxa.
We were encouraged by the capacity of eDNA collected from surface water samples to detect invasive O. rusticus in the exceptionally large Great Lakes, and suggest that this result may have relevance to eDNA applications for benthic arthropods in marine environments, as well (e.g., Kelly et al., 2016). Our six positive detections of O. rusticus eDNA within the Great Lakes all appear reasonable in light of their relatively close proximity to historic localities for this species reported by Peters et al. (2014). Similarly, four of our non-detections of O. rusticus eDNA occurred at sites where the nearest known historic locality for this crayfish was relatively remote; specifically, at the undeveloped Apostle Islands National Lakeshore, geographically isolated Beaver Island, and shorelines of the cities of Muskegon and Rochester, which lacked O. rusticus records nearby in lakes Michigan and Ontario, respectively (Fig. 1). However, eDNA samples from Duluth and Tawas Point did not detect O. rusticus eDNA, despite being close to localities for this species reported by Peters et al. (2014). In these two cases, O. rusticus records were from the years 1999 and 2002, and it is possible that O. rusticus could have experienced population declines or collapses at these sampling sites over the intervening timespan (Simberloff & Gibbons, 2004). Conversely, it is also possible that if O. rusticus is present but rare at these locations that our level of replication of eDNA sampling was not adequate to detect this crayfish, although Dougherty et al. (2016) found eDNA to be sensitive to detection of O. rusticus down to very low relative abundances in smaller inland lakes with similar replication. Future tests of the ability of eDNA to detect crayfishes in large freshwater habitats like the Great Lakes would benefit from pairing of this emerging methodology to concurrent, conventional sampling approaches like baited trapping or visual searches by divers or snorkelers (Larson & Olden, 2016).
eDNA detected P. leniusculus in six of seven lakes from which it was previously known to occur in our California and Nevada study region, and did not detect this crayfish from two lakes where it has never previously been observed. However, we did not detect P. leniusculus eDNA from one lake with a high previous estimate of relative abundance for this crayfish by baited trapping. Similar to our two potential errors of omission or false negatives for O. rusticus in the Great Lakes, this California lake had last been sampled for crayfish by baited trapping in a preceding year (2013), and a population decline or collapse could have occurred prior to our subsequent eDNA sampling. Yet a related issue is the apparently poor agreement between eDNA copy number and estimates of crayfish relative abundance by baited trapping observed both between and within our study lakes. In this case, timing of baited trapping is seemingly an inadequate explanation for disagreement between sampling methodologies, because at least within Lake Tahoe the majority of sites had been sampled for crayfish by baited trapping only a few days prior to our 2015 water collection for eDNA. Further, this result is consistent with Dougherty et al. (2016), who found little relationship between eDNA copy number and relative abundance of crayfish estimated by baited trapping that occurred immediately following water sample collection.
At present, eDNA copy number does not appear to be a good surrogate for crayfish relative abundance as estimated by baited trapping. This could be the result of biases or limitations associated with baited trapping for crayfish, including dependency of this passive sampling method on behavior (rather than exclusively abundance) of target organisms, which can be influenced by factors including the presence or abundance of crayfish predators (Collins et al., 1983; Larson & Olden, 2016). Alternatively, it could be because these comparisons to date have primarily used surface water samples for eDNA in comparison to trapping data for a benthic organism (but see Tréguier et al., 2014), and this might have been especially relevant in a large lake like Tahoe where P. leniusculus is known to use exceptionally deep water habitats (Abrahamsson & Goldman, 1970; Electronic Supplementary Table S4). Future tests of eDNA performance for crayfish or other benthic arthropods could benefit from taking water samples from deeper habitats over depth gradients to determine if the match between eDNA copy number and other estimates of relative abundance can be improved.
Our study contributes to a developing literature that increasingly suggests eDNA may be a viable monitoring tool for occupancy, if perhaps not abundance, of crayfish. Although Tréguier et al. (2014) found some disagreements between eDNA detections of the invasive red swamp crayfish Procambarus clarkii (Girard, 1852) in French ponds as related to results of baited trapping for this species, Dougherty et al. (2016) instead found generally high concordance between baited trapping estimates of presence or absence and eDNA results for O. rusticus from inland lakes of the northern US. Beyond applications to invasive crayfishes, Ikeda et al. (2016) tested an eDNA assay for the endangered crayfish Cambaroides japonicus (De Haan, 1841) in streams of Japan, and detected eDNA for this crayfish from all sites where it was manually collected. Improvements in performance of eDNA for detecting crayfish occupancy, or better reflecting relative abundance for these species, might also be achieved from more mechanistic laboratory studies that can address habitat or environmental factors affecting eDNA persistence times or transport distances (Barnes & Turner, 2016; Goldberg et al., 2016; Shogren et al., 2016). For example, Figiel & Bohn (2015) used laboratory studies to evaluate where crayfish eDNA was most prevalent, finding most frequent detections for the species Procambarus zonangulus (Hobbs & Hobbs, 1990) in sediments rather than surface waters, similar to results for some fish species (Turner et al., 2015). Ultimately, crayfish are both highly imperiled globally (Richman et al., 2015) and have also produced several major invasive species (Lodge et al., 2012a), and eDNA offers promising applications for better monitoring and management of both rare and introduced crayfishes.
eDNA is rapidly moving from proof of concept to actual implementation for purposes like surveillance for the early arrival of new or spreading invasive species (e.g., Jerde et al., 2013; Egan et al., 2015) or documenting distributional or population trends for imperiled species (e.g., de Souza et al., 2016). Here we report the first application of eDNA to test for major range expansions of invasive crayfishes into new regions where they were not previously known to occur, but where climatic conditions are anticipated to be suitable for their population establishment and spread (Larson & Olden, 2012; Morehouse & Tobler, 2013). Although we did not detect O. rusticus from California and Nevada or P. leniusculus from the Great Lakes, our ability to detect each crayfish from its reciprocal, known invasive range supports that eDNA can likely be used in surveillance for new invasions by benthic arthropods into even large aquatic habitats like the Great Lakes. In addition, our study involved successful collaboration with citizen scientists to collect some of our eDNA water samples, making it among the first efforts to combine citizen science with eDNA sampling for monitoring of biodiversity (Biggs et al., 2015; Miralles et al., 2016). For example, unsupervised citizen scientists collected water samples that detected O. rusticus eDNA from Cleveland on Lake Erie, yet no citizen scientist water samples were affected by contamination or false positives that might be a concern for merging these two sampling methodologies. Together, our study provides an optimistic demonstration for eDNA to be applied in biodiversity monitoring for increasingly diverse taxa and habitats by increasingly diverse communities of researchers.
References
Abrahamsson, S. A. & C. R. Goldman, 1970. Distribution, density and production of the crayfish Pacifastacus leniusculus Dana in Lake Tahoe, California-Nevada. Oikos 21: 83–91.
Barnes, M. A. & C. R. Turner, 2016. The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics 17: 1–17.
Barnes, M. A., C. R. Turner, C. L. Jerde, M. A. Renshaw, W. L. Chadderton & D. M. Lodge, 2014. Environmental conditions influence eDNA persistence in aquatic systems. Environmental Science and Technology 48: 1819–1827.
Biggs, J., N. Ewald, A. Valentini, C. Gaboriaud, T. Dejean, R. A. Griffiths, J. Foster, J. W. Wilkinson, A. Arnell, P. Brotherton & P. Williams, 2015. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biological Conservation 183: 19–28.
Capelli, G. M. & J. J. Magnuson, 1983. Morphoedaphic and biogeographic analysis of crayfish distribution in northern Wisconsin. Journal of Crustacean Biology 3: 548–564.
Capinha, C., B. Leung & P. Anastácio, 2011. Predicting worldwide invasiveness for four major problematic decapods: an evaluation of using different calibration sets. Ecography 34: 448–459.
Capinha, C., L. Brotons & P. Anastácio, 2013. Geographical variability in propagule pressure and climatic suitability explain the European distribution of two highly invasive crayfish. Journal of Biogeography 40: 548–558.
Collins, N. C., H. H. Harvey, A. J. Tierney & D. W. Dunham, 1983. Influence of predatory fish density on trapability of crayfish in Ontario lakes. Canadian Journal of Fisheries and Aquatic Sciences 40: 1820–1828.
de Souza, L. S., J. C. Godwin, M. A. Renshaw & E. Larson, 2016. Environmental DNA (eDNA) detection probability is influenced by seasonal activity of organisms. PLoS ONE 11: e0165273.
Doi, H., R. Inui, Y. Akamatsu, K. Kanno, H. Yamanaka, T. Takahara & T. Minamoto, 2016. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshwater Biology. doi:10.1111/fwb.12846.
Dougherty, M. M., E. R. Larson, M. A. Renshaw, C. A. Gantz, S. P. Egan, D. M. Erickson & D. M. Lodge, 2016. Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. Journal of Applied Ecology 53: 722–732.
Edwards, B. A., D. A. Jackson & K. M. Somers, 2009. Multispecies crayfish declines in lakes: implications for species distributions and richness. Journal of the North American Benthological Society 28: 719–732.
Egan, S. P., E. Grey, B. Olds, J. L. Feder, S. T. Ruggiero, C. E. Tanner & D. M. Lodge, 2015. Rapid molecular detection of invasive species in ballast and harbor water by integrating environmental DNA and light transmission spectroscopy. Environmental Science and Technology 49: 4113–4121.
Ficetola, G. F., C. Miaud, F. Pompanon & P. Taberlet, 2008. Species detection using environmental DNA from water samples. Biology Letters 4: 423–425.
Figiel, C. R. & S. Bohn, 2015. Laboratory experiments for the detection of environmental DNA of crayfish: examining the potential. Freshwater Crayfish 21: 159–164.
Gallardo, B., M. Clavero, M. I. Sánchez & M. Vilà, 2016. Global ecological impacts of invasive species in aquatic ecosystems. Global Change Biology 22: 151–163.
Goldberg, C. S., D. S. Pilliod, R. S. Arkle & L. P. Waits, 2011. Molecular detection of vertebrates in stream water: a demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE 6: e22746.
Goldberg, C. S., C. R. Turner, K. Deiner, K. E. Klymus, P. F. Thomsen, M. A. Murphy, S. F. Spear, A. McKee, S. J. Oyler-McCance, R. S. Cornman & M. B. Laramie, 2016. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution 7: 1299–1307.
Gunawardana, M., S. Chang, A. Jimenez, D. Holland-Moritz, H. Holland-Moritz, T. P. La Val, C. Lund, M. Mullen, J. Olsen, T. A. Sztain, J. Yoo, J. A. Moss & M. M. Baum, 2014. Isolation of PCR quality microbial community DNA from heavily contaminated environments. Journal of Microbiological Methods 102: 1–7.
Ikeda, K., H. Doi, K. Tanaka, T. Kawai & J. N. Negishi, 2016. Using environmental DNA to detect an endangered crayfish Cambaroides japonicus in streams. Conservation Genetics Resources 8: 231–234.
Jerde, C. L., W. L. Chadderton, A. R. Mahon, M. A. Renshaw, J. Corush, M. L. Budny, S. Mysorekar & D. M. Lodge, 2013. Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Canadian Journal of Fisheries and Aquatic Sciences 70: 522–526.
Kelly, R. P., J. L. O’Donnell, N. C. Lowell, A. O. Shelton, J. F. Samhouri, S. M. Hennessey, B. E. Feist & G. D. Williams, 2016. Genetic signatures of ecological diversity along an urbanization gradient. PeerJ 4: e2444.
Larson, E. R. & J. D. Olden, 2012. Using avatar species to model the potential distribution of emerging invaders. Global Ecology and Biogeography 21: 1114–1125.
Larson, E. R. & J. D. Olden, 2013. Crayfish occupancy and abundance in lakes of the Pacific Northwest, USA. Freshwater Science 32: 94–107.
Larson, E. R. & J. D. Olden, 2016. Field sampling techniques for crayfish. In Longshaw, M. & P. Stebbing (eds), Biology and Ecology of Crayfish. CRC Press, Boca Raton: 287–323.
Larson, E. R. & B. W. Williams, 2015. Historical biogeography of Pacifastacus crayfishes and their branchiobdellan and entocytherid ectosymbionts in western North America. In Kawai, T., Z. Faulkes & G. Scholtz (eds), Freshwater Crayfish: A Global Overview. CRC Press, Boca Raton: 404–447.
Larson, E. R., C. L. Abbott, N. Usio, N. Azuma, K. A. Wood, L.-M. Herborg & J. D. Olden, 2012. The signal crayfish is not a single species: cryptic diversity and invasions in the Pacific Northwest range of Pacifastacus leniusculus. Freshwater Biology 57: 1823–1838.
Larson, E. R., M. Castelin, B. W. Williams, J. D. Olden & C. L. Abbott, 2016. Phylogenetic species delimitation for crayfishes of the genus Pacifastacus. PeerJ 4: e1915.
Lodge, D. M., A. Deines, F. Gherardi, D. C. Yeo, T. Arcella, A. K. Baldridge, M. A. Barnes, W. L. Chadderton, J. L. Feder, C. A. Gantz & G. W. Howard, 2012a. Global introductions of crayfishes: evaluating the impact of species invasions on ecosystem services. Annual Review of Ecology, Evolution, and Systematics 43: 449–472.
Lodge, D. M., C. R. Turner, C. L. Jerde, M. A. Barnes, W. L. Chadderton, S. P. Egan, J. L. Feder, A. R. Mahon & M. E. Pfrender, 2012b. Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA. Molecular Ecology 21: 2555–2558.
Lodge, D. M., P. W. Simonin, S. W. Burgiel, R. P. Keller, J. M. Bossenbroek, C. L. Jerde, A. M. Kramer, E. S. Rutherford, M. A. Barnes, M. E. Wittmann, W. L. Chadderton, J. L. Aprisenig, D. Beletsky, R. M. Cooke, J. M. Drake, S. P. Egan, D. C. Finnoff, C. A. Gantz, E. K. Grey, M. H. Hoff, J. G. Howeth, R. A. Jensen, E. R. Larson, N. E. Mandrak, D. M. Mason, F. A. Martinez, T. J. Newcomb, J. D. Rothlisberger, A. J. Tucker, T. W. Warziniack & H. Zhang, 2016. Risk analysis and bioeconomics of invasive species to inform policy and management. Annual Review of Environment and Resources 41: 453–488.
Longmire, J. L., M. Maltbie & R. J. Baker, 1997. Use of “Lysis Buffer” in DNA Isolation and Its Implication for Museum Collections. Museum of Texas Tech University.
Matsuhashi, S., H. Doi, A. Fujiwara, S. Watanabe & T. Minamoto, 2016. Evaluation of the environmental DNA method for estimating distribution and biomass of submerged aquatic plants. PLoS ONE 11: e0156217.
Miralles, L., E. Dopico, F. Devlo-Delva & E. Garcia-Vazquez, 2016. Controlling populations of invasive pygmy mussel (Xenostrobus securis) through citizen science and environmental DNA. Marine Pollution Bulletin 110: 127–132.
Morehouse, R. L. & M. Tobler, 2013. Invasion of rusty crayfish, Orconectes rusticus, in the United States: niche shifts and potential future distribution. Journal of Crustacean Biology 33: 293–300.
Olden, J. D., J. W. Adams & E. R. Larson, 2009. First record of Orconectes rusticus (Girard, 1852) (Decapoda, Cambaridae) west of the great continental divide in North America. Crustaceana 82: 1347–1351.
Peters, J. A., M. J. Cooper, S. M. Creque, M. S. Kornis, J. T. Maxted, W. L. Perry, F. W. Schueler, T. P. Simon, C. A. Taylor, R. F. Thoma, D. G. Uzarski & D. M. Lodge, 2014. Historical changes and current status of crayfish diversity and distribution in the Laurentian Great Lakes. Journal of Great Lakes Research 40: 35–46.
Renshaw, M. A., B. P. Olds, C. L. Jerde, M. M. McVeigh & D. M. Lodge, 2015. The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol–chloroform–isoamyl alcohol DNA extraction. Molecular Ecology Resources 15: 168–176.
Richman, N. I., M. Böhm, S. B. Adams, F. Alvarez, E. A. Bergey, J. J. Bunn, Q. Burnham, J. Cordeiro, J. Coughran, K. A. Crandall, K. L. Dawkins, R. J. DiStefano, N. E. Doran, L. Edsman, A. G. Eversole, L. Füreder, J. M. Furse, F. Gherardi, P. Hamr, D. M. Holdich, P. Horwitz, K. Johnston, C. M. Jones, J. P. G. Jones, R. L. Jones, T. G. Jones, T. Kawai, S. Lawler, M. López-Mejía, R. M. Miller, C. Pedraza-Lara, J. D. Reynolds, A. M. M. Richardson, M. B. Schultz, G. A. Schuster, P. J. Sibley, C. Souty-Grosset, C. A. Taylor, R. F. Thoma, J. Walls, T. S. Walsh & B. Collen, 2015. Multiple drivers of decline in the global status of freshwater crayfish (Decapoda: Astacidea). Philosophical Transactions of the Royal Society B. doi:10.1098/rstb.2014.0060.
Roussel, J. M., J. M. Paillisson, A. Tréguier & E. Petit, 2015. The downside of eDNA as a survey tool in water bodies. Journal of Applied Ecology 52: 823–826.
Shogren, A. J., J. L. Tank, E. A. Andruszkiewicz, B. Olds, C. Jerde & D. Bolster, 2016. Modelling the transport of environmental DNA through a porous substrate using continuous flow-through column experiments. Journal of the Royal Society Interface 13: 20160290.
Simberloff, D., 2003. How much information on population biology is needed to manage introduced species? Conservation Biology 17: 83–92.
Simberloff, D. & L. Gibbons, 2004. Now you see them, now you don’t! Population crashes of established introduced species. Biological Invasions 6: 161–172.
Smart, A. S., R. Tingley, A. R. Weeks, A. R. van Rooyen & M. A. McCarthy, 2015. Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecological Applications 25: 1944–1952.
Svec, D., A. Tichopad, V. Novosadova, M. W. Pfaffl & M. Kubista, 2015. How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomolecular Detection and Quantification 3: 9–16.
Tréguier, A., J. M. Paillisson, T. Dejean, A. Valentini, M. A. Schlaepfer & J. M. Roussel, 2014. Environmental DNA surveillance for invertebrate species: advantages and technical limitations to detect invasive crayfish Procambarus clarkii in freshwater ponds. Journal of Applied Ecology 51: 871–879.
Tucker, A. J., W. L. Chadderton, C. L. Jerde, M. A. Renshaw, K. Uy, C. Gantz, A. R. Mahon, A. Bowen, T. Strakosh, J. M. Bossenbroek, J. L. Sieracki, D. Beletsky, J. Bergner & D. M. Lodge, 2016. A sensitive environmental DNA (eDNA) assay leads to new insights on Ruffe (Gymnocephalus cernua) spread in North America. Biological Invasions 18: 3205–3222.
Turner, C. R., K. L. Uy & R. C. Everhart, 2015. Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biological Conservation 183: 93–102.
Twardochleb, L. A., J. D. Olden & E. R. Larson, 2013. A global meta-analysis of the ecological impacts of nonnative crayfish. Freshwater Science 32: 1367–1382.
Untergasser, A., I. Cutcutache, T. Koressaar, J. Ye, B. C. Faircloth, M. Remm & S. G. Rozen, 2012. Primer3 – new capabilities and interfaces. Nucleic Acids Research 40: e115.
Usio, N., N. Azuma, E. R. Larson, C. L. Abbott, J. D. Olden, H. Akanuma, K. Takamura & N. Takamura, 2016. Phylogeographic insights into the invasion history and secondary spread of the signal crayfish in Japan. Ecology and Evolution 6: 5366–5382.
Vander Zanden, M. J., G. J. Hansen, S. N. Higgins & M. S. Kornis, 2010. A pound of prevention, plus a pound of cure: early detection and eradication of invasive species in the Laurentian Great Lakes. Journal of Great Lakes Research 36: 199–205.
Wegleitner, B. J., C. L. Jerde, A. Tucker, W. L. Chadderton & A. R. Mahon, 2015. Long duration, room temperature preservation of filtered eDNA samples. Conservation Genetics Resources 7: 789–791.
Acknowledgements
We are grateful to Anne Schrimpf and Thomas Horvath for providing tissue samples of European crayfishes of the family Astacidae used in primer testing for P. leniusculus, as well as to all zoo and aquarium staff and volunteers who assisted with collection of our eDNA water samples throughout the Great Lakes region. Our manuscript was improved by comments from two anonymous reviewers. This research was supported by US Environmental Protection Agency Grant EPA-R5-GL2012-1 to the University of Notre Dame and US Department of Agriculture National Institute of Food and Agriculture Hatch Project 1008988 to the University of Illinois.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: Koen Martens, Sidinei M. Thomaz, Diego Fontaneto and Luigi Naselli-Flores / Emerging Trends in Aquatic Ecology II
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Larson, E.R., Renshaw, M.A., Gantz, C.A. et al. Environmental DNA (eDNA) detects the invasive crayfishes Orconectes rusticus and Pacifastacus leniusculus in large lakes of North America. Hydrobiologia 800, 173–185 (2017). https://doi.org/10.1007/s10750-017-3210-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-017-3210-7