Abstract
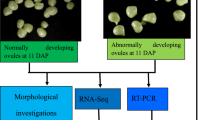
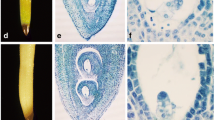
In hazelnuts, ovule abortion is associated with the formation of empty nuts, resulting in serious yield losses. Phytohormones play important roles in plant growth and development; however, the mechanisms through which they regulate ovule abortion in hazelnut are largely unknown. RNA-seq identified 1209 differentially expressed genes with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation between developing and abortive ovules in hazelnut. Hormone signal transduction (ko04075), and several phytohormone biosynthesis pathways, were found to be significantly enriched after pathway enrichment analysis. Compared with developing ovules, the levels of indole-3-acetic acid (IAA), gibberellins (GA), cytokinin (CTK), and salicylic acid (SA) were found to be significantly lower in abortive ovules, while levels of abscisic acid (ABA), ethylene (ETH), and jasmonic acid (JA) were found to be significantly higher. Consistent with these results, some important rate-limiting enzymes involved in the biosynthesis of phytohormones, including 9-cis-epoxycarotenoid dioxygenase, ACC synthase, and phospholipase A1 were significantly up-regulated in abortive ovules. Some key genes in the ETH, JA, and ABA signal transduction pathways were highly upregulated, including ethylene-responsive transcription factor 1 (ERF1/2) and the serine/threonine-protein kinase CTR1 in the ETH signal pathway, jasmonate ZIM domain-containing protein (JAZ) and the transcription factor MYC2 in the JA signal pathway, and ABA responsive element binding factor (ABF) in the JA signal pathway. Thus, ABA, ETH, and JA signaling were more active in abortive ovules than in developing ovules. Expression levels of NCED, ABF, ACS, ERF1/2, PLA1, and MYC2 revealed by q-RT-PCR were consistent with the results obtained by differentially expressed genes (DEG) analysis. Our data suggest that multiple phytohormone signaling pathways are involved in the development of hazelnut ovules. Inhibition of IAA, GA, CTK, and SA biosynthesis and signaling pathways, and activation of ABA, ETH, and JA biosynthesis and signaling pathways may contribute to the formation of abortive ovules in hazelnut.
Similar content being viewed by others
Abbreviations
- IAA:
-
Indole-3-acetic acid
- GA:
-
Gibberellins
- CTK:
-
Cytokinin
- ETH:
-
Ethylene
- ABA:
-
Abscisic acid
- JA:
-
Jasmonic acid
- BR:
-
Brassinolide
- SA:
-
Salicylic acid
- DEG:
-
Differentially expressed genes
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78. doi:10.1105/tpc.006130
Ariizumi T, Lawrence PK and Steber CM (2011) The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis GA signaling. Plant Physiol 155: 765–775. doi:10.1104/pp.110.166272
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7:986–995. doi:10.1101/gr.7.10.986
Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314:1410–1411. doi:10.1126/science.1134040
Bosco R, Caser M, Ghione GG, Mansuino A, Giovannini A, Scariot V (2015) Dynamics of abscisic acid and indole-3-acetic acid during the early-middle stage of seed development in rosa, hybrida. Plant Growth Regul 75:265–270. doi:10.1007/s10725-014-9950-8
Bradford MA, Reynolds JF (2008) Soil carbon stocks in experimental mesocosms are dependent on the rate of labile carbon, nitrogen and phosphorus inputs to soils. Funct Ecol 22:964–974. doi:10.1111/j.1365-2435.2008.01404.x
Chen Z, Zheng Z, Huang J, Lai Z, Fan B (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav 4:493–496. doi:10.4161/psb.4.6.8392
Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues. Arabidopsis Genes Dev 20:1790–1799. doi:10.1101/gad.1415106
Cheng Y, Qin G, Dai X, Zhao Y (2007) NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis. Arabidopsis P Natl Acad Sci USA 104:18825–18829. doi:10.1073/pnas.0708506104
Cheng YQ, Liu JF, Yang XD, Ma R, Liu Q, Liu CM (2013a) Construction of ethylene regulatory network based on the phytohormones related gene transcriptome profiling and prediction of transcription factor activities in soybean. Acta Physiol Plant 35(4):1303–1317. doi:10.1007/s11738-012-1170-0
Cheng Y, Liu J, Yang X, Ma R, Liu Q, Liu C (2013b) Construction of ethylene regulatory network based on the phytohormones related gene transcriptome profiling and prediction of transcription factor activities in soybean. Acta Physiol Plant 35:1303–1317. doi:10.1007/s11738-012-1170-0
Cheng YQ, Liu JF, Yang X, Ma R, Liu C, Liu Q (2013c) RNA-seq analysis reveals ethylene-mediated reproductive organ development and abscission in soybean (Glycine max L. Merr.). Plant Mol Biol Rep 31:607–619. doi:10.1007/s11105-012-0533-4
Cheng YQ, Liu JF, Zhang HD, Wang J, Zhao YX, Geng WT (2015) Transcriptome analysis and gene expression profiling of abortive and developing ovules during fruit development in hazelnut. Plos One 10:e0122072. doi:10.1371/journal.pone.0122072
Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, LorenzoO, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448:666–671. doi:10.1038/nature06006
Clouse SD (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23:1219–1230. doi:10.1105/tpc.111.084475
Gao XH, Xiao SL, Yao QF, Wang YJ, Fu XD (2011) An updated GA signaling ‘relief of repression’ regulatory model. Mol Plant 4:601–606. doi:10.1093/mp/ssr046
He HY, He LF, Gu MH, Li XF (2012) Nitric oxide improves aluminum tolerance by regulating hormonal equilibrium in the root apices of rye and wheat. Plant Sci 183:123–130. doi:10.1016/j.plantsci.2011.07.012
Hedden P, Phillip AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5:523–530. doi:10.1016/S1360-1385(00)01790-8
Hong JK, Lee SC, Hwang BK (2005) Activation of pepper basic PR-1 gene promoter during defense signaling to pathogen, abiotic and environmental stresses. Gene 356:169–180. doi:10.1016/j.gene.2005.04.030
Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19:884–894. doi:10.1016/j.devcel.2010.10.024
Hu XW, Huang XH, Wang YR (2012) Hormonal and temperature regulation of seed dormancy and germination. Leymus Chinensis Plant Growth Regul 67:199–207. doi:10.1007/s10725-012-9677-3
Huang XQ, He RQ, Liao XY, Zhou B, Peng WS, Lin JZ, Tang DY, Zhu YH, Zhao XY, Liu XM (2014) Effect of exogenous gibberellin on reserve accumulation during the seed filling stage of oilseed rape. Genet Mol Res 13:2827–2839. doi:10.4238/2014.January.22.7
Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24:1695–1708. doi:10.1101/gad.1953910
Jürgens G (2001) Apical–basal pattern formation in Arabidopsis embryogenesis. EMBO J 20:3609–3616. doi:10.1093/emboj/20.14.3609
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. doi:10.1093/nar/gkm882
Kemal K, Manners JM (2013) MYc2: the master in action. Mol Plant 6:686–703. doi:10.1093/mp/sss128
Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72:427–441. doi:10.1016/0092-8674(93)90119-B
Laurie-Berry N, Joardar V, Street IH, Kunkel BN (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 19:789–800. doi:10.1094/MPMI-19-0789
Liu S (2014) Cloning and expression analysis of cDNAs encoding ABA 8′-hydroxylase in peanut plants in response to osmotic stress. Plos One 9:e97025. doi:10.1371/journal.pone.0097025
Liu JF, Cheng YQ, Yan K, Liu Q, Wang ZW (2012) The relationship between reproductive growth and blank fruit formation in Corylus heterophylla fisch. Sci Hortic 136:128–134. doi:10.1016/j.scienta.2012.01.008
Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Joseph RE, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction. Arabidopsis Plant Cell 17:3007–3018. doi:10.1105/tpc.105.035451
Müller B, Sheen J (2008) Cytokinin and auxin interplay in root stem-cell specification during early embryogenesis. Nature 453:1094–1097. doi:10.1038/nature06943
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185. doi:10.1146/annurev.arplant.56.032604.144046
Nickstadt A, Thomma BP, Feussner IVO, Kangasjärvi J, Zeier J, Loeffler C, Scheel D, Berger S (2004) The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol Plant Pathol 5:425–434. doi:10.1111/j.1364-3703.2004.00242.x
Normanly J (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. CSH Perspect Biol 2:118–126. doi:10.1101/cshperspect.a001594
Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16:3216–3229. doi:10.1105/tpc.104.025387
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi:10.1038/nprot.2008.73
Singh DP, Jermakow AM, Swain SM (2002) Gibberellins are required for seed development and pollen tube growth. Arabidopsis Plant cell 14:3133–3147. doi:10.1105/tpc.003046
Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12:3703–3714. doi:10.1101/gad.12.23.3703
Sun X, Shantharaj D, Kang X, Ni M (2010) Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr Opin Plant Biol 13:611–620. doi:10.1016/j.pbi.2010.08.009
Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35:44–56. doi:10.1046/j.1365-313X.2003.01786.x
Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14:301–319. doi:10.1105/tpc.010283
Acknowledgments
This work was funded by the following grants: National Natural Science Foundation of China (31370683), Science and Technology Project of Jilin Province (20140101145JC), and Science and technology research project of The Education Department of Jilin Province (2016228).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cheng, Y., Zhao, Y., Liu, J. et al. Comparison of phytohormone biosynthesis and signal transduction pathways in developing and abortive hazelnut ovules. Plant Growth Regul 81, 147–157 (2017). https://doi.org/10.1007/s10725-016-0196-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0196-5