Abstract
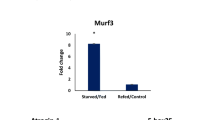
Autophagy is an important evolutionary conserved process in eukaryotic organisms for the turnover of intracellular substances. Recent studies revealed that autophagy displays circadian rhythms in mice and zebrafish. To date, there is no report focused on the rhythmic changes of autophagy in fish skeletal muscles upon nutritional deprivation. In this study, we examined the circadian rhythms of 158 functional genes in tilapia muscle in response to starvation. We found that 12 genes were involved in autophagy changed their rhythm after starvation. Among these genes, Atg4c, Bnip3la, Lc3a, Lc3b, Lc3c, and Ulk1a exhibited a daily rhythmicity in tilapia muscle, and Atg4b, becn1, bnip3la, bnip3lb, Lc3a, and ulk1b were significantly upregulated in response to starvation. The number of autophagosomes was dramatically increased in fasted fish, indicating that nutritional signals affect not only the muscular clock system but also its autophagy behavior. Administration of GSK4112, an activator of Nr1d1, altered rhythmic expression of both circadian clock genes and autophagy genes in tilapia muscle. Taken together, these findings provide evidence that nutritional deficiency affects both circadian regulation and autophagy activities in skeletal muscle.








Similar content being viewed by others
References
Allali KE et al. (2017) The Suprachiasmatic nucleus of the dromedary camel (Camelus dromedarius): cytoarchitecture and neurochemical anatomy Front Neuroanat 11:103
Bhadra U, Thakkar N, Das P, Pal BM (2017) Evolution of circadian rhythms: from bacteria to human Sleep Med:49–61
Bujak AL et al (2015) AMPK activation of muscle autophagy prevents fasting-induced hypoglycemia and myopathy during aging. Cell Metab 21:883–890
Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM (2012) Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485:123–127
Huang J, Zhong Z, Wang M, Chen X, Tan Y, Zhang S, He W, He X, Huang G, Lu H, Wu P, Che Y, Yan YL, Postlethwait JH, Chen W, Wang H (2015) Circadian modulation of dopamine levels and dopaminergic neuron development contributes to attention deficiency and hyperactive behavior. J Neurosci 35:2572–2587
Huang G, Zhang F, Ye Q, Wang H (2016) The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Rev-erbalpha and indirectly via Cebpb/(C/ebpbeta) in zebrafish. Autophagy 12:1292–1309
Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR (2008) Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci U S A 105:14946–14951
Lazado CC, Kumaratunga HP, Nagasawa K, Babiak I, Giannetto A, Fernandes JM (2014) Daily rhythmicity of clock gene transcripts in atlantic cod fast skeletal muscle PLoS One 9
Ma D, Panda S, Lin JD (2011) Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. Embo J 30:4642–4651
Mazzoccoli G et al (2013) Circadian transcriptome analysis in human fibroblasts from Hunter syndrome and impact of iduronate-2-sulfatase treatment. BMC Med Genomics 6:1755–8794
McCarthy JJ, Andrews JL, McDearmon E, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA (2007) Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics 31:86–95
Ojha R, Singh SK, Bhattacharyya S, Dhanda RS, Rakha A, Mandal AK, Jha V (2014) Inhibition of grade dependent autophagy in urothelial carcinoma increases cell death under nutritional limiting condition and potentiates the cytotoxicity of chemotherapeutic agent. J Urol 191:1889–1898
Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23:1860–1873
Reznick J, Preston E, Wilks DL, Beale SM, Turner N, Cooney GJ (2013) Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Biochim Biophys Acta 1:228–238
Seiliez I et al (2010) An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Com Biochem Physiol B Biochem Mol Biol 157:258–266
Toledo M et al (2018) Autophagy regulates the liver clock and glucose metabolism by degrading CRY1. Cell Metab 28:S1550413118303309
Wang M, Zhong Z, Zhong Y, Zhang W, Wang H (2015) The zebrafish period2 protein positively regulates the circadian clock through mediation of retinoic acid receptor (RAR)-related orphan receptor alpha (Roralpha). J Biol Chem 290:4367–4382
Wier AM et al (2010) Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A 107:2259–2264
Wu P et al (2016) Daily rhythmicity of clock gene transcript levels in fast and slow muscle fibers from Chinese perch (Siniperca chuatsi). BMC Genomics 17:016–3373
Wu P et al. (2018a) Impact of short-term fasting on the rhythmic expression of the core circadian clock and clock-controlled genes in skeletal muscle of crucian carp (Carassius auratus) Genes 9
Wu P, Chu W, Liu X, Guo X, Zhang J (2018b) The influence of short-term fasting on muscle growth and fiber hypotrophy regulated by the rhythmic expression of clock genes and myogenic factors in Nile tilapia. Mar Biotechnol 20:750–768
Xiong X, Tao R, Depinho RA, Dong XC (2012) The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem 287:39107–39114
Acknowledgments
We thank the National Natural Science Foundation of China for supporting the funding and Collaborative Innovation Center for Efficient and Health Production of Fisheries, Hunan Province for their support and assistance with the fish.
Funding
This study was supported by the National Natural Science Foundation of China (No. 31820103016). The funding bodies did not have a role in the design of the study, data collection, analysis, interpretation of data, writing the manuscript, nor the decision to publish.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that there are no conflicts of interest.
Consent for publication
This is not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, P., Cheng, J., Chen, L. et al. Nr1d1 affects autophagy in the skeletal muscles of juvenile Nile tilapia by regulating the rhythmic expression of autophagy-related genes. Fish Physiol Biochem 46, 891–907 (2020). https://doi.org/10.1007/s10695-019-00757-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-019-00757-9