Abstract
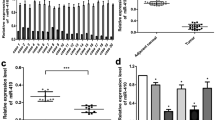
Gliomas are the most common and the most malignant brain tumors, accouting for 45–55 % of all intracranial tumors. The incidence of glioma worldwide is about 6–12 per 100,000. Recently, several studies showed that the activation of the oncogenes and the inactivation and/or loss of the tumor suppressor genes, especially for miRNA-21, let-7 and so on, are the most primary molecule event in gliomas. MicroRNAs (miRNAs) are a class of endogenously expressed small noncoding RNAs which are usually 21–23 nucleotides long. miRNAs regulate gene expression and play important roles in a variety of physiological and pathological processes, such as cell proliferation, differentiation and apoptosis. To date, Growing evidence has shown that mi RNAs are frequently dysregulated in human cancers and can act as both tumor suppressors and oncogenes. Along with the discovery of micro RNA, more and more research focusing on its relationship with glioma was carried out to investigate the biological features of glioma and to provide experimental evidence for glioma mechanism. In the present study, we aimed to verify the miRNA-126 down-regulation which showed in the results of glioma tissue miRNAs chip and discuss the miRNA-126 methylation in patients with glioma. A total of 50 samples from patients with glioma and 20 control samples from patients with cerebral trauma were included in this study. The expression levels of the miR-126 gene were detected using quantitative polymerase chain reaction (PCR), and the methylation status of miR-126 was examined using methylation-specific PCR-denaturing high-performance liquid chromatography (MSP–DHPLC). The expression level of miRNA-126 was found to be significantly higher in the control group (0.6134 ± 0.1214) than in the glioma group (0.2771 ± 0.1529; P < 0.05). The expression was also significantly elevated in low-grade gliomas (0.3117 ± 0.1474) compared with high-grade gliomas (0.1582 ± 0.1345; P < 0.05). In addition, increased methylation of miR-126 was found in 40 % of glioma patients in our study (20/50 cases), resulting in significantly decreased miR-126 expression (0.1715 ± 0.1376; P < 0.05). Our results indicate that we verified successfully the miRNA-126 down-regulation phenomenon in patients with glioma which showed in the results of glioma tissue miRNAs chip and the miRNA-126 down-regulation through methylation in patients with glioma. So we could say that epigenetic modification is a crucial mechanism for controlling the expression of miR-126 in glioma.

Similar content being viewed by others
References
Di Stefano AL, Enciso-Mora V, Marie Y, Desestret V, Labussiere M, Boisselier B et al (2013) Association between glioma susceptibility loci and tumour pathology defines specific molecular etiologies. Neuro Oncol 15:542–547
He LW, Shi R, Jiang L, Zeng Y, Ma WL, Zhou JY (2014) XRCC1 gene polymorphisms and glioma risk in Chinese population: a meta-analysis. PLoS One 9:e111981
Huse JT, Holland EC (2010) Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer 10:319–331
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507
Yu J, Cai X, He J, Zhao W, Wang Q, Liu B (2012) Microarray-based analysis of gene regulation by transcription factors and microRNAs in glioma. Neurol Sci 34:1283–1289
Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G et al (2005) Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334:1351–1358
Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS et al (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28:5369–5380
Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M et al (2008) miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 6:14
Hansen TF, Nielsen BS, Jakobsen A, Sorensen FB (2015) Intra-tumoural vessel area estimated by expression of epidermal growth factor-like domain 7 and microRNA-126 in primary tumours and metastases of patients with colorectal cancer: a descriptive study. J Transl Med 13:10
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD et al (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451:147–152
Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ et al (2008) The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun 377:136–140
Ebrahimi F, Gopalan V, Smith RA, Lam AK (2013) miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol 96:98–107
Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11:241–247
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14:1902–1910
Kim YK, Kim VN (2007) Processing of intronic microRNAs. EMBO J 26:775–783
Smith ZD, Meissner A (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14:204–220
Kishi M, Nakamura M, Nishimine M, Ikuta M, Kirita T, Konishi N (2005) Genetic and epigenetic alteration profiles for multiple genes in salivary gland carcinomas. Oral Oncol 41:161–169
Nakahara Y, Shintani S, Mihara M, Hino S, Hamakawa H (2006) Detection of p16 promoter methylation in the serum of oral cancer patients. Int J Oral Maxillofac Surg 35:362–365
Moreira PR, Guimaraes MM, Guimaraes AL, Diniz MG, Gomes CC, Brito JA et al (2009) Methylation of P16, P21, P27, RB1 and P53 genes in odontogenic keratocysts. J Oral Pathol Med 38:99–103
Mirimanoff RO (2014) High-grade gliomas: reality and hopes. Chin J Cancer 33:1–3
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol 4:199–227
Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10:94–108
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114
Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79:351–379
Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23:175–205
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Han L, Yue X, Zhou X, Lan FM, You G, Zhang W et al (2012) MicroRNA-21 expression is regulated by beta-catenin/STAT3 pathway and promotes glioma cell invasion by direct targeting RECK. CNS Neurosci Ther 18:573–583
Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65:6029–6033
Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH et al (2009) MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem 23:347–358
Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M et al (2009) MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun 380:205–210
Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S et al (2009) microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res 1269:158–165
Jiang L, Mao P, Song L, Wu J, Huang J, Lin C et al (2010) miR-182 as a prognostic marker for glioma progression and patient survival. Am J Pathol 177:29–38
Yu F, Zheng J, Mao Y, Dong P, Li G, Lu Z et al (2015) Long non-coding RNA APTR promotes the activation of hepatic stellate cells and the progression of liver fibrosis. Biochem Biophys Res Commun 463:679–685
Xiong Y, Kotian S, Zeiger MA, Zhang L, Kebebew E (2015) miR-126-3p inhibits thyroid cancer cell growth and metastasis, and is associated with aggressive thyroid cancer. PLoS One 10:e0130496
Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N et al (2014) Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget 5:11873–11885
Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G (2009) Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun 379:726–731
Funding information
The research was supported by the college and university research program of the education department of Inner Mongolia Autonomous Region (No. NJZY13420).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Rights and permissions
About this article
Cite this article
Cui, H., Mu, Y., Yu, L. et al. Methylation of the miR-126 gene associated with glioma progression. Familial Cancer 15, 317–324 (2016). https://doi.org/10.1007/s10689-015-9846-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-015-9846-4