Abstract
The melon aphid Aphis gossypii Glover is one of the most serious pests in cucumber production, often causing severe damage in commercial fields. Identifying and deploying resistant germplasm and understanding the inheritance of melon aphid resistance are required for cucumber geneticists to develop an effective breeding strategy. In this study, resistance of 30 cucumber selections to melon aphid was evaluated at the seedling stage. Six generations, P1, P2, F1, BC1, BC2 and F2 derived from the cross of JY30 (susceptible) × EP6392 (resistant), were used as genetic populations to study the inheritance of melon aphid resistance in cucumber with the mixed major gene plus polygene inheritance model with the joint analysis method of multiple generations. Eight of the 30 tested selections displayed resistance to the melon aphid. The resistance of cucumber to melon aphids was controlled by one additive and dominant major gene plus additive and dominant polygenes, and was affected by environment as well. The additive effect and the dominant effect of the major gene were greater than the additive effect and the dominant effect of the polygenes. The heritabilities of the major gene in BC1, BC2 and F2 were 63.62 %, 0 % and 70.39 %, respectively. The polygenic heritabilities were 22.62 %, 37.0 % and 9.32 %, and the ratios of the environmental variance to phenotype variance were 58.54 %, 63.16 % and 30.77 %. We conclude that selections of cucumber with high resistance to melon aphid could be screened in advanced generations.
Similar content being viewed by others
Introduction
Cucumber (Cucumis sativus L.) belongs to the family Cucurbitaceae (Jeffrey 1980; Mliki et al. 2003) and is one of the most important vegetable crops in many countries of the world. The melon aphid (Aphis gossypii Glover) has occurred in most areas of China. It often causes severe damage and is one of the most serious pests in cucumber production. Identifying and deploying resistant germplasm and understanding the inheritance of melon aphid resistance in cucumbers is crucial for breeding aphid-resistant cucumbers with high quality.
Proper assessment of plant resistance is necessary for successful research on plant aphid resistance. It was reported that the aphid damage exponent could be used as the basis for identifying aphid resistance in cotton (Zhang and Yuan 1978) and alfalfa (Wu et al. 2007). The average number of aphids per plant was used as a basis for identifying aphid resistance in bean (Han et al. 1991). There is evidence for genetic control of aphid resistance in many crops, including tomato (Solanum lycopersicum) (Rossi et al. 1998), wheat (Triticum aestivum) (Liu et al. 2005), barley (Hordeum vulgare) (Mittal et al. 2008), melon (C. melo) (Sarria et al. 2008; Brotman et al. 2002), barrel clover (Medicago truncatula) (Klingler et al. 2005), maize (Zea mays) (So et al. 2010) and soybean (Glycine max) (Kim et al. 2010a, b; Ohnishi et al. 2012). However, little is known about the inheritance of aphid resistance in cucumber.
It is assumed that in addition to the major gene, a polygenic component is included in the inheritance system for quantitative traits. Such a mixed major gene plus polygene inheritance model is derived from the mixed one major gene plus polygene inheritance theory according to Elston (1984). This theory accounts for some inheritance of quantitative traits that could not be explained by classical Mendelian genetics. The mixed major gene plus polygene genetic model has been widely used in genetic analysis of multiple traits in rice (Gai and Wang 1998), wheat (Zhang et al. 2007a), soybean (Wang and Gai 2001; Li et al. 2008) and rape (Zhang et al. 2006). This model is also applied to the genetic analysis of multiple traits of cucumber, including downy mildew resistance (Zhang et al. 2007b) and fruit stalk length (Ma et al. 2010). However, this model has not been used in the study of the inheritance of cucumber aphid resistance.
In the present study, identification of the differences among 30 cucumber selections to melon aphid resistance was carried out based on the numbers of aphids per seedling. JY30 and EP6392 were chosen as susceptible and resistant aphid parents to produce F1, F2, BC1 and BC2 generations for genetic analysis following a mixed inheritance model with the objective of elucidating the inheritance of aphid resistance in cucumber.
Materials and methods
Plant materials and aphid infection
The aphid resistance level of 30 cucumber selections, including ZaoerN, NingYang7, CaiLv2, A’Xin, JY30, RiJieCheng, PingWang cucumber, BaiGuiFei, BiYu, EP6392, WanLv, JIN5-508, XUE1, D8, JinPengBaiYu, GY14A, ZaoKang, Katya, DDX, EP326, RiYin2, ZhongNong115, EP6411, YuNv, JinZhou cucumber, MaPiHuang, Burpee, GY2, CiXi cucumber and XiaoYe, was evaluated by counting the number of aphids per cucumber seedling. Seeds of each selection were sown in trays filled with potting substrate (nutrient available: 40–60 g/kg total NPK nutrients, ≥350 g/kg total humus content, 6.5–7.5 pH) in three environmental growth chambers maintained at 25 °C (18 h)/18 °C (6 h) day/night, light intensity of 12,000 lux (18 h)/0 lux (6 h), and relative humidity ranged from 50 to 60 % on 2 December 2012. Seven days after the germination, five apterous adult melon aphids were transferred to the back of the first true leaf per seedling. Ten plants of each selection were infested with aphids, and three replications were conducted.
JY30 (P1, susceptible female parent), and EP6392 (P2, resistant male parent) were used to produce F1 progeny based on the number of aphids per plant and plant reaction trait during March and June 2011. Selfing F1 plants produced an F2 population, and F1 was crossed with P1 to produce BC1 and F1 was crossed with P2 to produce BC2 during March and June 2013. Experiments were performed at the Yangzhou University Department of Horticulture research farm. Seeds of P1, P2, F1, F2, BC1 and BC2 generations were sown in trays filled with potting substrate (nutrient available: 40–60 g/kg total NPK nutrients, ≥350 g/kg total humus content, 6.5–7.5 pH) in four environmental growth chambers maintained at 25 °C (18 h)/18 °C (6 h) day/night, light intensity of 12,000 lux (18 h)/0 lux (6 h), and relative humidity ranged from 50 to 60 % on 6 July 2013. The number of sown seeds of P1, P2, F1, F2, BC1 and BC2 were 60, 60, 60, 440, 100 and 100, respectively. Seven days after the germination, five apterous adult melon aphids were transferred to the back of the first true leaf per seedling.
Determination of aphid resistance
The numbers of aphids on individual plants were counted at 8 days after aphid infestation by assigning aphid scores ranging from 1 to 5, where 1 ≤ 100 aphids per plant, 2 = 101–200 aphids per plant, 3 = 201–300 aphids per plant, 4 = 301–400 aphids per plant and 5 ≥ 401 aphids per plant, modified from Jun et al. (2012).
Data analysis
Data from six generations were used for inheritance analysis by the joint segregation analysis (JSA) method (Gai and Wang 1998). The principle of JSA is as follows: first, we assumed that the segregating population was composed of component distributions controlled by a major gene and was modified by both polygenes and the environment. A total of five groups and 24 types of genetic models were established, including a “one major gene inheritance” model (A), a “two major genes inheritance” model (B), a “polygenic inheritance” model (C), a “mixed one major gene and one polygene inheritance” model (D) and a “mixed two major genes and a polygene inheritance” model (E). Subsequently, a joint maximum-likelihood function was derived using data from six generations to estimate the parameters of component distributions through the iterated expectation and conditional maximization (IECM) algorithm. Additionally, the Akaike’s information criterion (AIC), likelihood-ratio test (LRT), and a set of goodness-of-fit tests were used for model selection and testing. The model with the least AIC value and best fitness was considered as the best-fitting model. Related genetic parameters, including gene effects and genetic variances of major genes and polygenes were obtained from estimates of component distributions. Finally, individuals from segregating populations were classified into major-gene genotypes according to their posterior probabilities (Gai and Wang 1998; Zhang et al. 2000; Gai et al. 2003; Yao et al. 2013).The AIC value and related genetic parameters were calculated using the Segreg Anal Soft Program (http://jpkc.njau.edu.cn/swtj/show.asp?classid=35&articleid=44&classtype=26) and Statistical Analysis System (SAS, Cary, NC, USA).
Results
Evaluation of cucumber aphid resistance
Screening germplasm for aphid resistance led to the discovery of aphid-resistant selections within the 30 tested cucumber selections (Table 1). We identified eight aphid-resistant cucumber selections, including Katya, EP326, JIN5-508, Burpee, ZaoKang, EP6392, EP6411 and GY14A, as well as two highly susceptible selections, JY30 and BaiGuiFei.
The leaves of aphid-resistant cucumber selections were curled slightly upwards, whereas those of aphid-susceptible selections were more obviously curled upwards. Plants with aphid resistance performed normally in growth and development, but plants with aphid susceptibility were severely stunted at 8 day after aphid infestation.
Variation of aphid resistance among the six generations
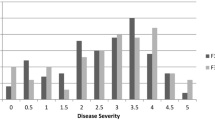
‘EP6392’ showed significant resistance to aphid infestation, whereas ‘JY30’was susceptible (Table 2). The F1 population had a tendency toward the resistant parent; BC1 and BC2 showed a single mode in the susceptible direction and resistant direction, respectively; and the F2 population showed two modes in both the susceptible and resistant directions (Table 2).
Inheritance analysis
To estimate the number of genes and heritability parameters, the aphid scores of individual plants of six generations were analyzed using the Segreg Anal Soft Program. The maximum log likelihood value (MLV), AIC value and the maximum-likelihood estimates in each genetic model were calculated. The model D-4 had the smallest AIC value of 1508.00, followed by E-1 (1536.07) (Table 3). Therefore, D-4 and E-1 were chosen as candidates for a best-fitting model to explain the inheritance of aphid resistance in ‘JY30’ × ‘EP6392’ progenies according to Akaike’s hypothesis on maximizing expected entropy (Akaike 1977).
The tests for goodness-of-fit between the expected values from the selected model and the observed values were conducted to determine whether the selected model sufficiently explained the data. LRT and test of goodness-of-fit for the models D-4 and E-1 indicated that model D-4 was the best-fitting genetic model to explain the inheritance of aphid resistance in the cross (Table 4). This means that aphid resistance in the cross of ‘JY30 × EP6392’ was controlled by one additive and dominant major gene plus additive and dominant polygenes.
The results of genetic components of each segregated generation showed that the additive effect and the dominant effect of the major gene were greater than the additive effect of the polygenes (Table 5). The dominant degrees of the major gene and polygenes were about −1.00 and −0.8. The heritability of the major gene in BC1, BC2 and F2 was 63.62 %, 0.00 % and 70.39 %, respectively. The polygenic heritability was 22.62 %, 37.00 % and 9.32 %, respectively, and the ratios of the environmental variance to phenotype variance was 58.54 %, 63.16 % and 30.77 %, respectively. These results indicated that the environmental effect was also important in cucumber resistance to melon aphids.
Discussion
The melon aphid is one of the major pests in cucumber production. It feeds specifically from the sieve element and causes damage by removing plant nutrients and carbohydrates. Severe aphid infestation may cause many visible symptoms, including curling, wilting, yellowing and plant stunting. Use of aphid-resistant selections increases yields and improves cucumber quality, and also protects the environment by allowing reduced insecticide application. Identification of germplasm with aphid resistance is an effective way to explore the crop’s genetic resources; however, proper assessment is necessary. The ratio of aphid number was used as the basis for identifying aphid resistance in chrysanthemum (He et al. 2010). Aphid damage index was used as a basis for identifying aphid resistance in soybean (Meng et al. 2010), cotton (Zhang and Yuan 1978) and alfalfa (Wu et al. 2007). In this study, an assessment of cucumber aphid resistance was conducted by assigning aphid scores according to the number of aphids per seedling in chambers. The results showed that eight of the 30 tested selections were resistant to the melon aphid. Our results may provide aphid-resistant materials for the study of genetic mechanisms of aphid resistance in cucumber and for breeding work.
Previous studies have shown that inheritance of aphid resistance may be monogenic or polygenic. For example, inheritance of greenbug resistance is controlled by a single dominant resistance gene (Gb3) in wheat (Weng and Lazar 2002; Weng et al. 2005; Azhaguvel et al. 2012). Mi was also found to confer resistance to the potato aphid (Macrosiphum euphorbiae) and whitefly (Bemisia tabaci) and the Mi-1 gene was cloned (Milligan et al. 1998; Rossi et al. 1998; Vos et al. 1998). A dominant gene (Dp-1) provides resistance to the pear aphid (Dysaphis pyri) (Evans et al. 2008). Three dominant genes, Rag1, Rag2 and Rag3, for resistance to the soybean aphid (A. glycines) were mapped to independent soybean linkage groups (Hill et al. 2009; Zhang et al. 2010). Four quantitative trait loci (QTLs) and two pairs of epistatic QTLs for resistance to melon-cotton aphid were mapped, and the major gene vat gene was cloned (Boissot et al. 2010; Pauquet et al. 2004). Different species might have different inheritance models for aphid resistance. The present results showed that genetic model D-4 was the best fitting genetic model for melon aphid resistance in cucumber. It indicated that melon aphid resistance in cucumber was controlled by one additive and dominant major gene plus additive and dominant polygenes. The heritability of the major gene and polygenes in F2 was about 70.39 and 9.32 %. This result showed that cucumber selections with aphid resistance could be screened in advanced generations. The environmental and other variances in F2 were approximately 20.29 %. This indicated that aphid resistance in cucumber could be enhanced by improving cultivation conditions and optimizing cultivation techniques.
References
Akaike H (1977) On the entropy maximum principle. In: Krishnaiah PR (ed) Applications of statistics. North-Holland Publishing Company, Amsterdam, pp 27–41
Azhaguvel P, Rudd JC, Ma YQ, Luo MC, Weng YQ (2012) Fine genetic mapping of greenbug aphid-resistance gene Gb3 in Aegilops tauschii. Theor Appl Genet 124:555–564
Boissot N, Thomas S, Sauvion N, Marchal C, Pavis C, Dogimont C (2010) Mapping and validation of QTLs for resistance to aphids and whiteflies in melon. Theor Appl Genet 121:9–20
Brotman Y, Silberstein L, Kovalski I et al (2002) Resistance gene homologues in melon are linked to genetic loci conferring disease and pest resistance. Theor Appl Genet 104:1055–1063
Elston RC (1984) The genetic analysis of quantitative trait differences between two homozygous lines. Genetics 108:733–744
Evans KM, Govan CL, Fernandez-Fernandez F (2008) A new gene for resistance to Dysaphis pyri in pear and identification of flanking microsatellite markers. Genome 51:1026–1031
Gai JY, Wang JK (1998) Identification and estimation of a QTL model and its effects. Theor Appl Genet 97:1162–1168
Gai JY, Zhang YM, Wang JK (2003) Genetic system of quantitative traits in plants. Science Press, Beijing
Han WZ, Cao J, Wang XL, Cao RH (1991) Identification of bean germplasm resources resistant to bean aphid. China Seeds 1:32–33
He JP, Chen FD, Chen SM, Fang WM (2010) Aphid-resistance of chrysanthemum cultivars. Chin J Ecol 29:1382–1386
Hill CB, Kim KS, Crull L, Diers BW, Hartman GL (2009) Inheritance of resistance to the soybean aphid in soybean PI 200538. Crop Sci 49:1193–1200
Jeffrey C (1980) A review of the Cucurbitaceae. Bot J Linn Soc 81:223–247
Jun TH, Mian MAR, Michel AP (2012) Genetic mapping revealed two loci for soybean aphid resistance in PI 567301B. Theor Appl Genet 124:13–22
Kim KS, Bellendir S, Hudson KA et al (2010a) Fine mapping the soybean aphid resistance gene Rag1 in soybean. Theor Appl Genet 120:1063–1071
Kim KS, Hill CB, Hartman GL, Hyten DL, Hudson ME, Diers BW (2010b) Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theor Appl Genet 121:599–610
Klingler J, Creasy R, Gao LL et al (2005) Aphid resistance in Medicago truncatula involves antixenosis and phloem-specific, inducible antibiosis, and maps to a single locus flanked by NBS-LRR resistance gene analogs. Plant Physiol 137:1445–1455
Li GJ, Cheng LG, Zhang GZ, He XH, Zhi HJ, Zhang YM (2008) Mixed major-gene plus polygenes inheritance analysis for resistance in soybean to bean pyralid (Lamprosema indicata Fabricius). Soybean Sci 27:33–41
Liu XM, Smith CM, Friebe BR, Gill BS (2005) Molecular mapping and allelic relationships of Russian wheat aphid-resistance genes. Crop Sci 45:2273–2280
Ma J, Si LT, Tian Y (2010) Mixed major gene and polygene inheritance analysis of fruit stalk length in cucumber. Acta Agric Boreali-Occidentalis Sin 19:161–165
Meng FL, Li WB, Duan YX, Zhang DY, Li DM, Wang ZK (2010) Identification techniques and screening of soybean aphid resistant germplasm. Soybean Sci 29:457–460
Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10:1307–1319
Mittal S, Dahleen LS, Mornhinweg D (2008) Locations of quantitative trait loci conferring Russian wheat aphid resistance in barley germplasm STARS-9301B. Crop Sci 48:1452–1458
Mliki A, Staub JE, Sun ZY, Ghorbel A (2003) Genetic diversity in African cucumber (Cucumis sativus L.) provides potential for germplasm enhancement. Genet Resour Crop Evol 50:461–468
Ohnishi S, Miyake N, Takeuchi T et al (2012) Fine mapping of foxglove aphid (Aulacorthum solani) resistance gene Raso1 in soybean and its effect on tolerance to Soybean dwarf virus transmitted by foxglove aphid. Breed Sci 61:618–624
Pauquet J, Burget E, Hagen L et al (2004) Map-based cloning of the Vat gene from melon conferring resistance to both aphid colonization and aphid transmission of several viruses. Progress in cucurbit genetics and breeding research. In: Proceedings of Cucurbitaceae 2004, the 8th EUCARPIA meeting on cucurbit genetics and breeding, Olomouc, Czech Republic, 12–17 July, p 325–329
Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95:9750–9754
Sarria E, Yuste-Lisbona FJ, Palomares FJ, López-Sesé AI, Gómez-Guillamón ML (2008) Inheritance of tolerance to Aphis gossypii in C. melo TGR-1551 and its relation with resistance to virus transmission. Cucurbitaceae 2008—IX EUCARPIA Meeting (poster session)
So YS, Ji HC, Brewbaker JL (2010) Resistance to corn leaf aphid (Rhopalosiphum maidis Fitch) in tropical corn (Zea mays L.). Euphytica 172:373–381
Vos P, Simons G, Jesse T et al (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol 16:1365–1369
Wang JK, Gai JY (2001) Mixed inheritance model for resistance to agromyzid beanfly (Melunugromyzu sojue Zehntner) in soybean. Euphytica 122:9–18
Weng YQ, Lazar MD (2002) Amplified fragment length polymorphism- and simple sequence repeat-based molecular tagging and mapping of greenbug resistance gene Gb3 in wheat. Plant Breed 121:218–223
Weng YQ, Li W, Devkota RN, Rudd JC (2005) Microsatellite markers associated with two Aegilops tauschii-derived greenbug resistance loci in wheat. Theor Appl Genet 110:462–469
Wu DG, He CG, Wu TJ, Tang SR, Jia B (2007) Resistance comparison of eleven alfalfa varieties to aphid. Grassl Turf (Bimonthly) 4:54–57
Yao MH, Li N, Wang F, Ye ZB (2013) Genetic analysis and identification of QTLs for resistance to cucumber mosaic virus in chili pepper (Capsicum annuum L.). Euphytica 193:135–145
Zhang JL, Yuan F (1978) The exponent method of aphid damage investigation and its practical application. J Northwest A&F Univ 54–62
Zhang YM, Gai JY, Wang JK (2000) Identification of two major genes plus polygenes mixed inheritance model of quantitative trait in B1 and B2, and F2. J Biomath 15:358–366
Zhang SF, Ma CZ, Zhu JC, Wang JP, Wen YC, Fu TD (2006) Genetic analysis of oil content in Brassica napus L. using mixed model of major gene and polygene. Acta Genet Sin 33:171–180
Zhang LP, Zhao CP, Shan FH et al (2007a) The mixed genetic analysis of photoperiod-temperature sensitive male sterility of BS219 in wheat. Acta Agron Sin 33:1553–1557
Zhang SQ, Gu XF, Zhang SP, Zou ZR (2007b) Inheritance of downy mildew resistance in cucumber (Cucumis sativus L.). Acta Bot Boreali-Occidentalia Sin 27:2416–2420
Zhang GR, Gu CH, Wang DH (2010) A novel locus for soybean aphid resistance. Theor Appl Genet 120:1183–1191
Acknowledgments
This research was financially supported by the National Program on Key Basic Research Projects (The 973 Program: 2012CB113900), Jiangsu Science & Technology Project (BE2012326) and the Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Liang, D., Hu, Q., Xu, Q. et al. Genetic inheritance analysis of melon aphid (Aphis gossypii Glover) resistance in cucumber (Cucumis sativus L.). Euphytica 205, 361–367 (2015). https://doi.org/10.1007/s10681-015-1391-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1391-6