Abstract
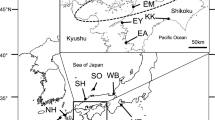
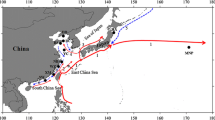
Identification of population units is crucial for management and monitoring programs, especially for endangered wild species. The roughskin sculpin (Trachidermus fasciatus Heckel) is a small catadromous fish and has been listed as a second class state protected aquatic animal since 1988 in China. To achieve sustainable conservation of this species, it is necessary to clarify the existing genetic structure both between and within populations. Here, population genetic structure among eight populations of T. fasciatus were analyzed by using 16 highly polymorphic microsatellites. High levels of genetic variation were observed in all populations. All pairwise F ST estimates were significant after false discovery rate correction (overall average F ST = 0.054). Furthermore, both STRUCTURE and discriminant analysis of principal components (DAPC) analysis showed that the eight populations were grouped into six clusters. BAYESASS analysis showed generally low recent and asymmetric migration among populations. All these results suggested significant genetic structure across populations. However, there was no isolation by distance relationship among populations, likely resulting from barriers to gene flow created by habitat fragmentation. Our results highlight the need for in situ conservation efforts for T. fasciatus across its entire distribution range, through maximizing habitat size and quality to preserve overall genetic diversity and evolutionary potential.



Similar content being viewed by others
References
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Allendorf FW, Luikart G (2009) Conservation and the genetics of populations. Wiley, New York
Balloux F, Brünner H, Lugon-Moulin N, Hausser J, Goudet J (2000) Microsatellites can be misleading: an empirical and simulation study. Evolution 54:1414–1422
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188
Besnier F, Glover KA (2013) ParallelStructure: A R package to distribute parallel runs of the population genetics program structure on multi-core computers. Plos One 8:e70651. doi:10.1371/journal.pone.0070651
Casu M, Rivera-Ingraham GA, Cossu P, Lai T, Sanna D, Dedola GL, Sussarellu R, Sella G, Cristo B, Curini-Galletti M, Garcia-Gomez JC, Espinosa F (2011) Patterns of spatial genetic structuring in the endangered limpet Patella ferruginea: implications for the conservation of a Mediterranean endemic. Genetica 139:1293–1308
Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989–2000
Daily GC, Luck GW, Ehrlich PR (2003) Population diversity and ecosystem services. Trends Ecol Evol 18:331–336
DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56:461–473
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellis J, Pashley C, McCAULEY D (2006) High genetic diversity in a rare and endangered sunflower as compared to a common congener. Mol Ecol 15:2345–2355
Ernest E, Haanes H, Bitanyi S, Fyumagwa R, Msoffe P, Bjørnstad G, Røed K (2012) Influence of habitat fragmentation on the genetic structure of large mammals: evidence for increased structuring of African buffalo (Syncerus caffer) within the Serengeti ecosystem. Conserv Genet 13:381–391
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Faubet P, Waples RS, Gaggiotti OE (2007) Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16:1149–1166
Frankham R (1995) Conservation genetics. Annu Rev Genet 29:305–327
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Frankham R, Briscoe DA, Ballou JD (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Gao TX, Bi XX, Zhao LL, Li CJ (2013) Population genetic structure of roughskin sculpin Trachidermus fasciatus based on the mitochondrial Cytb sequence. Acta Hydrobiol Sin 37:199–207 (in Chinese with English abstracts)
Garrigan D, Marsh PC, Dowling TE (2006) Long-term effective population size of three endangered colorado river fishes. Anim Conserv 5:95–102
Gerlach G, Musolf K (2000) Fragmentation of landscape as a cause for genetic subdivision in bank voles. Conserv Biol 14:1066–1074
Glenn TC, Schable NA (2005) Isolating microsatellite DNA loci. Methods Enzymol 395:202–222
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9. 3). Available from http://www2.unil.ch/popgen/softwares/fstat.htm
Hedrick PW (1999) Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 53:313–318
Hedrick PW (2004) Recent developments in conservation genetics. For Ecol Manag 197:3–19
Hilborn R, Quinn TP, Schindler DE, Rogers DE (2003) Biocomplexity and fisheries sustainability. Proc Natl Acad Sci USA 100:6564–6568
Hughes JB, Daily GC, Ehrlich PR (1997) Population diversity: its extent and extinction. Science 278:689–692
Islam MS, Hibino M, Tanaka M (2007) Distribution and diet of the roughskin sculpin, Trachidermus fasciatus, larvae and juveniles in the Chikugo River estuary, Ariake Bay, Japan. Ichthyol Res 54:160–167
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Jost L (2009) D versus GST: response to Heller and Siegismund (2009) and Ryman and Leimar (2009). Mol Ecol 18:2088–2091
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Li Y, Lancaster ML, Cooper SJB, Taylor AC, Carthew SM (2015) Population structure and gene flow in the endangered southern brown bandicoot (Isoodon obesulus obesulus) across a fragmented landscape. Conserv Genet 16:331–345
Liu JX, Avise JC (2011) High degree of multiple paternity in the viviparous Shiner Perch, Cymatogaster aggregata, a fish with long-term female sperm storage. Mar Biol 158:893–901
Ma ZT, Liu ZZ, Pan LD, Jiang X (2012) Genetic variation analysis of roughskin sculpin (Trachidermus fasciatus) in native populations of China by ISSR. J Fish China 36:1042–1048 (in Chinese with English abstracts)
Maes GE, Pujolar JM, Hellemans B, Volckaert FAM (2006) Evidence for isolation by time in the European eel (Anguilla anguilla L.). Mol Ecol 15:2095–2107
Mbora DN, McPeek MA (2010) Endangered species in small habitat patches can possess high genetic diversity: the case of the Tana River red colobus and mangabey. Conserv Genet 11:1725–1735
Meirmans PG (2014) Nonconvergence in bayesian estimation of migration rates. Mol Ecol Resour 14:726–733
Moore J, Miller H, Daugherty C, Nelson N (2008) Fine-scale genetic structure of a long-lived reptile reflects recent habitat modification. Mol Ecol 17:4630–4641
Moritz C (1999) Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas 130:217–228
Onikura N, Takeshita N, Matsui S, Kimura S (2002) Spawning grounds and nests of Trachidermus fasciatus (Cottidae) in the Kashima and Shiota estuaries system facing Ariake Bay, Japan. Ichthyol Res 49:198–201
Park SDE (2001) Trypanotolerance in West African cattle and the population genetic effects of selection. Dissertation, University of Dublin
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from https://www.R-project.org/
Røed KH, Bjørnstad G, Flagstad Ø, Haanes H, Hufthammer AK, Jordhøy P, Rosvold J (2014) Ancient DNA reveals prehistoric habitat fragmentation and recent domestic introgression into native wild reindeer. Conserv Genet 15:1137–1149
Rousset F (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Rousset F (2014) Genepop 4.3 for Windows/Linux/Mac OS X. This documentation: July, 8, 2014
Schmitt T, Seitz A (2002) Influence of habitat fragmentation on the genetic structure of Polyommatus coridon (Lepidoptera: Lycaenidae): implications for conservation. Biol Conserv 107:291–297
Shao BX, Tang ZY, Sun GY, Qiu YC, Shao YJ, Xue ZY (1980) On the breeding habitat of Trachidermus fasciatus Heckel. J Fish China 4:81–86 (in Chinese with English abstracts)
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Sremba AL, Hancock-Hanser B, Branch TA, LeDuc RL, Baker CS (2012) Circumpolar diversity and geographic differentiation of mtDNA in the critically endangered Antarctic blue whale (Balaenoptera musculus intermedia). Plos One. doi:10.1371/journal.pone.0032579
Tseng MC, Lee SC, Tzeng WN (2001) Genetic variation of the Japanese eel Anguilla japonica based on microsatellite DNA. J Taiwan Fish Res 9:137–147
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wang JQ (1999) Advances in studies on the ecology and reproductive biology of Trachidermus fasciatus Heckel. Acta Hydrobiol Sin 23:729–734 (in Chinese)
Wang YH (2006) Notes on the scientific name, type-locality and geographical distribution of roughskin sculpin (Trachidermis fasciatus Heckel, 1840). Mar Fish 4:005 (in Chinese with English abstracts)
Wang JQ, Cheng G (2010) The historical variance and causes of geographical distribution of a roughskin sculpin (Trachidermus fasciatus Heckel) in Chinese territory. Acta Ecol Sin 30:6845–6853 (in Chinese with English abstracts)
Wang JQ, Cheng G, Tang ZP (2001) The distribution of roughskin sculpin (Trachidermus fasciatus Heckel) in Yalu river basin, China. J Fudan Univ Nat Sci 40:471–476 (in Chinese with English abstracts)
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Yue PJ, Chen YY (1988) China red data book of endangered animals-pisces. Science Press, Beijing (in Chinese)
Acknowledgments
The study was supported by the National Natural Science Foundation of China (NSFC) (No. 31502169), the NSFC-Shandong Joint Fund for Marine Ecology and Environmental Sciences (No. U1406403), the National Marine Public Welfare Research Project (201405010), and 100 Talents Program of the Chinese Academy of Sciences to JXL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, YL., Xue, DX., Gao, TX. et al. Genetic diversity and population structure of the roughskin sculpin (Trachidermus fasciatus Heckel) inferred from microsatellite analyses: implications for its conservation and management. Conserv Genet 17, 921–930 (2016). https://doi.org/10.1007/s10592-016-0832-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-016-0832-7