Abstract
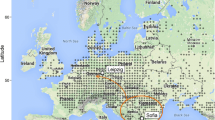
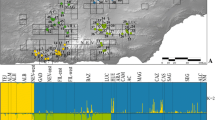
Genetic diversity is one of the most important criteria to identify unique populations for conservation purposes. In this study we analyze the genetic population structure of the endangered montane mayfly Ameletus inopinatus in its European range. The species is restricted to unpolluted cold-water streams, and exhibits an insular distribution across highlands of Central Europe and a more continuous distribution across Fennoscandia and Northern Euro-Siberia. We genotyped 389 individuals from 31 populations for eight highly polymorphic microsatellite loci to investigate genetic diversity and population structure within and among European mountain ranges. Genetic diversity of A. inopinatus decreases along an east–west gradient in Central Europe and along a north–south gradient in Fennoscandia, respectively. Centres of exceptionally high genetic diversity are located in the Eastern Alps (Andertal Moor, Austria), the High Tatra, the Beskides, the Sudety Mountains and the Eastern German Highlands. Species distribution modelling for 2080 projects major regional habitat loss, particularly in Central Europe mountain ranges. By relating these range shifts to our population genetic results, we identify conservation units primarily in Eastern Europe, that if preserved would maintain high levels of the present-day genetic diversity and continue to provide long-term suitable habitat under future climate warming scenarios.


Similar content being viewed by others
References
Alcamo J, Bouwman A, Edmonds J, Grübler A, Morita T, Sugandhy A (1995) An evaluation of the IPCC IS92 emission scenarios. In: Climate change 1994, radiative forcing of climate change and an evaluation of the IPCC IS92 emission scenarios. Cambridge University Press, Cambridge, pp 233–304
Alsos IG, Alm T, Normand S et al (2009) Past and future range shifts and loss of diversity in dwarf willow (Salix herbacea L.) inferred from genetics, fossils and modelling. Glob Ecol Biogeogr 18:223–239
Araújo MB, New M (2006) Ensemble forecasting of species distributions. Trends Ecol Evol 22:42–47
Beebee TJC (2007) Population structure and its implications for conservation of the great silver beetle Hydrophilus piceus in Britain. Freshw Biol 52:2101–2111
Böhmer J, Rawer-Jost C, Zenker A (2004) Multimetric assessment of data provided by water managers from Germany: assessment of several different types of stressors with macrozoobenthos communities. Hydrobiologia 516:215–228
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetic studies. Mol Ecol 13:3261–3273
Buffagni A, Cazzola M, López-Rodríguez MJ, Alba-Tercedor J, Armanini DG (2009) In: Schmidt-Kloiber A, Hering D (eds) Distribution and ecological preferences of European freshwater organisms. Volume 3—Ephemeroptera. Pensoft Publishers, Sofia, pp 1–254
Bunn SE, Hughes JM (1997) Dispersal and recruitment in streams: evidence from genetic studies. J N Am Benthol Soc 16:338–346
Chakraborty R, De Andrade M, Daiger SP, Budowle B (1992) Apparent heterozygote deficiencies observed in DNA typing data and their implications in forensic applications. Ann Hum Genet 56:45–57
Cordellier M, Pfenninger M (2008) Climate driven range dynamics of the freshwater limpet, Ancylus fluviatilis (Pulmonata, Basommatophora). J Biogeogr 35:1580–1592
Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295
Daking EE, Avise JC (2004) Microsatellite null alleles in parentage analysis. Heredity 93:504–509
Duelli P (1997) Biodiversity evaluation in agricultural landscapes: an approach at two different scales. Agric Ecosyst Environ 62:81–91
EU Commission (2010). http://ec.europa.eu/environment/nature/index_en.htm. Accessed 18 March 2010
European Environment Agency (2010) http://www.eea.europa.eu/de. Accessed 18 March 2010
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial-DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin version 3.11: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Felsenstein J (2008) PHYLIP (Phylogeny inference package) version 3.68. Available at http://evolution.genetics.washington.edu/phylip/software.html. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Fiedler PL, Jain SK (1992) Conservation biology: the theory and practice of nature conservation, preservation and management. Chapman and Hall, New York
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Freeland JR (2005) Molecular ecology. Wiley, Hoboken
GBIF-Sweden (2010) GBIF data portal. http://data.gbif.org. Accessed 18 March 2010
Gienapp P, Teplitsky C, Alho JS, Mills JA, Merilä J (2008) Climate change and evolution: disentangling environmental and genetic responses. Mol Ecol 17:167–178
Glaubitz JC (2004) CONVERT: a user friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310
Graf W, Schultz H, Janececk B (2004) Ökofaunistische Erhebungen und Bewertung im Natura 2000-Gebiet St. Lorenzener Hochmoor, Makrozoobenthos Endbericht
Graham CH, Ferrier S, Huettman F, Moritz C, Townsend Peterson A (2004) New developments in museum-based informatics and applications in biodiversity analysis. Trends Ecol Evol 19:497–503
Guillot G, Mortier F, Estoup A (2005) GENELAND: a computer package for landscape genetics. Mol Ecol Notes 5:712–715
Harte J, Ostling A, Green JL, Kinzig A (2004) Climate change and extinction risk. Nature. doi:10.1038/nature02718
Haybach A (2003) Zoogeographische Aspekte der Eintagsfliegenbesiedlung Deutschlands (Insecta, Ephemeroptera). Verhandlungen der westdeutschen Entomologentagung Düsseldorf 2002:187–209
Hendrey GR, Wright RF (1976) Acid precipitation in Norway: effects on aquatic fauna. J Gt Lakes Res 2:192–207
Hering D, Schmidt-Kloiber A, Murphy J, Lücke S, Zamora-Muñoz C, López Rodríguez MJ, Huber T, Graf W (2009) Potential impact of climate change on aquatic insects: a sensitivity analysis for European caddisflies (Trichoptera) based on distribution patterns and ecological preferences. Aquat Sci 71:3–14
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Hoelzel E (1967) Die Fauna des Hochmoores von St. Lorenzen in den Gurker Alpen. Carinthia 195–211
Hoffmann AA, Willi Y (2008) Detecting genetic responses to environmental change. Nat Rev Genet 9:421–432
Houghton J (2001) The science of global warming. Interdiscip Sci Rev 26:247–257
Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M (2008) Ecological consequences of genetic diversity. Ecol Lett 11:609–623
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comp Graph Stat 5:299–314
Jackson JK, Resh VH (1992) Variation in genetic structure among populations of the caddisfly Helicopsyche borealis from three streams in northern California, U.S.A. Freshw Biol 27:29–42
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Leberg PL (2002) Estimating allelic richness: effects of sample size and bottlenecks. Mol Ecol 11:2445–2449
Malzacher P, Jacob U, Haybach A, Reusch H (1998) Rote Liste der Eintagsfliegen (Ephemeroptera). In: Binot M, Bless R, Boye P, Gruttke H, Pretscher P (eds) Rote Liste gefährdeter Tiere Deutschlands, Bundesamt für Naturschutz (BfN). Bonn-Bad Godesberg, Germany, pp 264–267
McNeely JA, Miller KR, Reid WV, Mittermeier RA, Werner TB (1990) Conserving the world’s biological diversity. World Conservation Union, World Resources Institute, Conservation International, World Wildlife Fund—US, and the World Bank, Washington, DC
Moussalli A, Moritz C, Williams SE, Carnaval AC (2009) Variable responses of skinks to a common history of rainforest fluctuation: concordance between phylogeography and palaeo-distribution models. Mol Ecol 18:483–499
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
OTA (US Congress Office of Technology Assessment) (1987) Technologies to maintain biological diversity. US Government Printing Office, Washington, DC
Parmesan C, Yohe G (2003) A globally coherent fingerprint of climate change impacts across natural systems. Nature 421:37–42
Pauls SU, Lumbsch T, Haase P (2006) Phylogeography of the montane caddisfly Drusus discolor: evidence for multiple refugia and periglacial survival. Mol Ecol 15:2153–2169
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Raymond M, Rousset F (1995) GENEPOP (version 1.2) population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rosenberg DM, Resh VH (1993) Freshwater biomonitoring and benthic invertebrates. Chapman & Hall, New York
Russev B, Vidinova Y (1994) Distribution and ecology of the representatives of some families of order Ephemeroptera (Insecta) in Bulgaria. Lauterbornia 19:107–113
SAS Institute Inc. (2007) JMP user guide. SAS Institute Inc, Cary
Schultz H, Janecek B, Hess M, Reusch H, Graf W (2004) Das Makrozoobenthos des Natura 2000 Gebietes St. Lorenzener Hochmoor (Andertal, Kernten) unter besonderer Berücksichtigung der Libellenfauna (Insecta: Odonata). Carinthia II 196:343–358
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Soldán T, Enktaivan S, Godunko RJ (2009) Commented checklist of mayflies (Ephemeroptera) of Mongolia. Aquat Insects 31:653–670
Taberlet P (1998) Biodiversity at the intraspecific level: the comparative phylogeographic approach. J Biotechnol 64:91–100
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Theissinger K, Feldheim KA, Taubmann J, Seitz A, Pauls SU (2008) Isolation and characterization of 10 higly polymorphic di- and trinucleotide microsatellite markers in the mayfly Ameletus inopinatus (Ephemeroptera: Siphlonuridae). Mol Ecol Resour 8:1285–1287
Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC et al (2004) Extinction risk from climate change. Nature 427:145–147
Van Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wilcock HR, Bruford MW, Nichols RA, Hildrew AG (2007) Landscape, habitat characteristics and the genetic population structure of two caddisflies. Freshw Biol 52:1907–1929
Wright JF, Sutcliffe DW, Furse MT (eds) (2000) Assessing the biological quality of fresh waters: RIVPACS and other techniques. Freshwater Biological Association, Ambleside, Cumbria, UK
Acknowledgments
We are very grateful to Ralf Brettfeld, John Brittain, Michael Theobald, Hanno Voigt and André Wagner who supported this study by providing material, information, and help in the field. This study was financially supported by the German Research Foundation (DFG) grant HA 3431/2-1 awarded to PH and SUP, and HA 3431/2-2 awarded to PH and Alfred Seitz (Mainz), and the research funding programme ‘‘LOEWE—Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz’’ of Hesse’s Ministry of Higher Education, Research, and the Arts. KT and IL are supported by PhD scholarships of the Studienstiftung des deutschen Volkes (SdV). SUP gratefully acknowledges a German Academy of Sciences Leopoldina Postdoctoral Research Fellowship (BMBF-LPD 9901/8-169).
Author information
Authors and Affiliations
Corresponding authors
Additional information
In memory of Prof. Dr. Alfred Seitz.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Taubmann, J., Theissinger, K., Feldheim, K.A. et al. Modelling range shifts and assessing genetic diversity distribution of the montane aquatic mayfly Ameletus inopinatus in Europe under climate change scenarios. Conserv Genet 12, 503–515 (2011). https://doi.org/10.1007/s10592-010-0157-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0157-x