Abstract
The adaptive potential of a species to a changing environment and in disease defence is primarily based on genetic variation. Immune genes, such as genes of the major histocompatibility complex (MHC), may thereby be of particular importance. In marsupials, however, there is very little knowledge about natural levels and functional importance of MHC polymorphism, despite their key role in the mammalian evolution. In a previous study, we discovered remarkable differences in the MHC class II diversity between two species of mouse opossums (Gracilinanus microtarsus, Marmosops incanus) from the Brazilian Atlantic forest, which is one of the most endangered hotspots for biodiversity conservation. Since the main forces in generating MHC diversity are assumed to be pathogens, we investigated in this study gastrointestinal parasite burden and functional associations between the individual MHC constitution and parasite load. We tested two contrasting scenarios, which might explain differences in MHC diversity between species. We predicted that a species with low MHC diversity would either be under relaxed selection pressure by low parasite diversity (‘Evolutionary equilibrium’ scenario), or there was a recent loss in MHC diversity leading to a lack of resistance alleles and increased parasite burden (‘Unbalanced situation’ scenario). In both species it became apparent that the MHC class II is functionally important in defence against gastrointestinal helminths, which was shown here for the first time in marsupials. On the population level, parasite diversity did not markedly differ between the two host species. However, we did observe considerable differences in the individual parasite load (parasite prevalence and infection intensity): while M. incanus revealed low MHC DAB diversity and high parasite load, G. microtarsus showed a tenfold higher population wide MHC DAB diversity and lower parasite burden. These results support the second scenario of an unbalanced situation.
Similar content being viewed by others
Introduction
Pathogens represent very powerful agents of selection that have the potential to drive rapid changes in the genetic composition of natural host populations. In the coevolutionary host-pathogen interplay they are particularly important in maintaining host genetic variation (McCallum and Dobson 1995; Altizer et al. 2003, 2007). On the other hand, pathogens do exhibit the potential of triggering serious population declines once host genetic diversity is lost due to inbreeding or genetic drift. In such unbalanced situations, pathogens may even pose a severe extinction risk to small populations that have lost their ability to buffer environmental challenges (McCallum and Dobson 1995; Woodroffe 1999; Altizer et al. 2003, 2007). A number of studies showed indeed that low levels of genetic diversity are associated with a reduced immune reaction and high pathogen loads (e.g. Liersch and Schmid-Hempel 1998; Coltman et al. 1999; Meagher 1999; Cassinello et al. 2001; Dorman et al. 2004; Spielman et al. 2004).
In terms of pathogen defence there is a group of well studied immune genes that plays a critical role in triggering the vertebrate adaptive immune response: the major histocompatibility complex (MHC). The MHC encodes receptor molecules that recognise and bind antigens in order to present them to T-lymphocytes (Klein 1986). While MHC class I molecules mainly present peptides derived from the inside of viral or cancer-infested cells, MHC class II molecules primarily correspond to extracellular pathogens (e.g. bacteria, nematodes, cestodes, Klein and Horejsi 1997). One of the most typical attributes of MHC genes is the enormous polymorphism found in the majority of vertebrates studied to date, generated and maintained by balancing selection (Klein 1986; Hedrick 1994; Apanius et al. 1997; Sommer 2005). Selection processes are thought to lead to a diversification of MHC alleles driven by the necessity to recognise a wide array of pathogens (Hedrick 1994; Bernatchez and Landry 2003; Sommer 2005; Piertney and Oliver 2006). Thereby, MHC heterozygotes could exhibit a selective advantage over homozygote individuals, because they possess a larger number of MHC variants to recognize and defend parasite antigens (‘heterozygote advantage’, Doherty and Zinkernagel 1975). Or certain MHC alleles could be advantageous, whereby selection is frequency-dependent and varies over time and space (‘rare-allele-advantage’, Takahata and Nei 1990; Slade and McCallum 1992; Hedrick 2002). The diversification of MHC alleles is on the other hand constrained by the need to delete T-cells that react with self peptide–MHC combinations, leading to the assumption that natural selection favours an optimal and intermediate number of MHC alleles per individual (Wegner et al. 2003a; Kurtz et al. 2004). Due to their central role in disease defence the genes of the MHC are also particularly suited as a genetic basis for mate choice (Tregenza and Wedell 2000). MHC dissimilar mating preferences might act to increase the heterozygosity of the progeny (Zuk 1990) or to avoid inbreeding or genetic incompatibility (Brown and Erklund 1994).
In marsupials, the MHC has been poorly investigated so far, although this group represents a milestone in the evolution of mammals. Studies on captive or laboratory bred marsupials indicated low levels of MHC class II diversity, and very limited polymorphism was assumed to be a general feature of the marsupial MHC class II (Stone et al. 1996, 1998). However, there is a lack of knowledge about natural levels of marsupial MHC class II diversity, aside from our previous study (Meyer-Lucht et al. 2008) and two other recent investigations (Siddle et al. 2007b; Holland et al. 2008a, b). The latter studies reported considerable MHC class II diversity in the Australian brushtail possum (Trichosurus vulpecula, Holland et al. 2008a, b), but no MHC class II variation in Tasmanian devils (Sarcophilus harrisii, Siddle et al. 2007b). Interestingly, our study on free-ranging Neotropical marsupials revealed contradictory patterns of MHC class II diversity in the two investigated species as well. Diversity was high in the Brazilian gracile mouse opossum (Gracilinanus microtarsus, Wagner 1842) and congruent with levels of MHC II diversity described in numerous wild eutherian species (Meyer-Lucht et al. 2008). In contrast, MHC II diversity was rather low (five times lower) in the second species under study, the Gray slender mouse opossum (Marmosops incanus, Lund 1840).
The two investigated mouse opossum species are endemic to the Brazilian Atlantic forest, which is one of the most threatened biomes in the world (Myers et al. 2000). Once ranging almost continuously along the Brazilian coast and covering more than 1.5 million km2, the Atlantic forest nowadays comprises only 12% of its original extent (SOS Mata Atlântica and Instituto Nacional de Pesquisas Espaciais 2008; Ribeiro et al. 2009). Nevertheless, it still harbours an extraordinary concentration of endemic species (Myers et al. 2000). Based on these facts the region has been classified as one of the five most important biodiversity hotspots for conservation priorities (Myers et al. 2000). With ongoing expansion of human activities, however, the remaining Atlantic forest patches are continuously being fragmented and degraded (Tabarelli et al. 2004, 2005; Teixeira et al. 2009).
The two study species, G. microtarsus and M. incanus, feature similar ecological characteristics: they are nocturnal, solitary (Caceres 2004) and feed on a similar omnivorous diet (Fonseca and Kierulff 1989; Martins and Bonato 2004). However, they differ in microhabitat preferences as G. microtarsus is mainly arboreal while M. incanus more frequently uses the understorey and forest ground (Cunha and Vieira 2002; Vieira and Monteiro-Filho 2003). Both species show at least partially a semelparous mating behaviour, which means that individuals contribute to only one mating season and die thereafter (Lorini et al. 1994; Martins et al. 2006).
In view of this apparent similar environmental background the question arises what causes the conspicuous differences in MHC diversity between the two mouse opossum species. Assuming that the main selective forces in generating and maintaining a high MHC diversity are pathogens (Apanius et al. 1997; Hughes and Yeager 1998), we focussed in this study on the parasite load of the two mouse opossum species, more precisely on the burden with gastrointestinal helminths. We considered two possible scenarios with respect to MHC diversity and parasite burden and predicted the following:
-
(1)
‘Evolutionary equilibrium’ scenario: at a state of evolutionary equilibrium between hosts and pathogens, selection through diverse pathogens will have caused high MHC polymorphism in a population or a species, whereas low MHC polymorphism indicates the presence of a less diverse pathogenic selection pressure. This scenario holds true if hosts and pathogens share a long-term coevolutionary history. This scenario was indeed supported by a comparison of different human populations worldwide (Prugnolle et al. 2005), and a study on stickle back populations from different habitats (Wegner et al. 2003b). Both investigations revealed that populations exposed to a more diverse pathogen regime exhibit higher MHC diversity than those exposed to fewer pathogens. And more recently, in a meta-analysis on rodent species and their helminth parasites, Goüy de Bellocq et al. (2008) detected likewise a significant positive correlation between MHC allelic diversity in a species and the number of different parasite species, the so called parasite diversity. According to this scenario, we expected to observe relative high parasite diversity in the MHC class II diverse species G. microtarsus, and relative low parasite diversity in the species with low MHC class II diversity, M. incanus.
-
(2)
‘Unbalanced situation’ scenario: as a contrasting scenario we assumed an unbalanced situation, which might be caused by a recent loss of genetic diversity in the depleted host by means of, for instance, a bottleneck event. The species with low MHC diversity could have lost resistance alleles or other important parts of its adaptive evolutionary potential. Moreover, after a recent loss the host population most likely is genetically more or less homogenous. This fact facilitates an easy spread of pathogens throughout the population, because most individuals share the same resistance genotype (Meagher 1999). According to this second scenario, we expected that the MHC depleted species should face higher parasite loads than the MHC diverse species, especially in terms of individual parasite prevalence and infection intensity.
We tested these theoretical assumptions by analysing the patterns of gastrointestinal helminth burden and MHC class II diversity in the two mouse opossum species G. microtarsus and M. incanus. In addition, we investigated functional associations between individual parasite load and MHC variation to understand selection processes acting on the marsupial MHC DAB under natural conditions.
Methods
Study area and sampling
The study was carried out in the Brazilian Atlantic forest in the region of Caucaia do Alto, municipalities of Cotia and Ibiúna, State of São Paulo, Brazil. Small mammal trapping was conducted in five forest fragments (sites S1 to S5) within a fragmented landscape with 31% native forest cover and one control site (CS) within an adjacent continuous forest at the Morro Grande Forest Reserve. We used Sherman live traps in a grid of 100 trap locations in 20 m intervals and captured in five trapping sessions per site from July 2003 to March 2005. Within this period, 102 Brazilian gracile mouse opossums (G. microtarsus) and 123 Gray slender mouse opossums (M. incanus) were trapped. G. microtarsus was not captured in forest site S2. From the trapped animals small ear cuts were taken for tissue samples (~5 mm2) and stored in 70% ethanol. Faeces were collected from the used traps and stored in 96% ethanol. Traps were cleaned prior to re-use. For detailed information on trapping sessions and study sites see Püttker et al. (2006), for further details upon the study area see Pardini et al. (2005).
Molecular analyses
We genotyped the MHC DAB genes of all 102 G. microtarsus and 123 M. incanus individuals to assess allelic diversity, and thereby doubled the sample size from our previous work (Meyer-Lucht et al. 2008). Genomic DNA from ear tissue samples was extracted using the DNeasy Tissue Kit (Qiagen, Hilden, Germany). A 195 bp fragment of the marsupial MHC class II DAB was amplified. This fragment codes for the major part of the β1 domain of the molecule and contains positively selected amino acid sites that are presumably involved in antigen binding (Meyer-Lucht et al. 2008). In the human DR1 molecule this part comprises a major part of the antigen binding sites as defined by Brown et al. (1993).
We used the forward primer JS1 (Schad et al. 2004) and the reverse primer ML8 (Meyer-Lucht et al. 2008) for PCR amplification. Single stranded conformation polymorphism (SSCP) analyses on 15% polyacrylamide gels were used to genotype M. incanus. SSCP band patterns were assigned to alleles, and exemplary bands were excised and re-amplified. Two Main-DAB alleles (Main-DAB*03 and Main-DAB*04) could not be distinguished on the SSCP patterns, and were therefore handled together as Main-DAB*03/04. The two alleles differed in one synonymous and one non-synonymous nucleotide substitution (Meyer-Lucht et al. 2008).
Due to the large number of alleles per individual G. microtarsus was genotyped via molecular cloning and sequencing of fifteen recombinant clones per individual. This number was determined from initial trials on extensive sequencing of recombinant clones in two individuals (Meyer-Lucht et al. 2008). Transformed clones were PCR amplified using the vector primers T7 and M13. Amplification products were sequenced on an ABI PRISM 310 Automated Genetic Analyzer (Applied Biosystems, Foster City, Ca, USA) using the BigDye Terminator v3.1 Cycle Sequencing Kit (ABI). A precise description of the molecular techniques is given in Meyer-Lucht et al. (2008). To ensure the comparability between the two quantification methods (SSCP and molecular cloning) applied for the two species, we performed both techniques in parallel for a subset of ten M. incanus individuals. The results are congruent to a large extent. Slight differences in the number of detected alleles did not affect our overall conclusions supporting the comparability between the two techniques in our study (Supplement Table 1).
Parasitological examinations
To analyse the degree of gastrointestinal helminth parasitism in G. microtarsus and M. incanus, presence and number of helminth eggs was non-invasively assessed in the host faecal samples. Faecal egg counts (FEC) are not only informative in terms of prevalence but also appropriate to assess the intensity of infections (Soulsby 1982). These counts reflect the overall worm burden and worm fecundity, which both are influenced by the immune state of the host (Stear et al. 1995, 1997). Due to its non-invasive nature FEC is applicable even to wildlife populations, hence, it is a widely used approach in field studies (e.g. Coltman et al. 1999; Cassinello et al. 2001; Ferrari et al. 2004; Seivwright et al. 2004; Froeschke and Sommer 2005; Harf and Sommer 2005; Meyer-Lucht and Sommer 2005; Schad et al. 2005; Schwensow et al. 2007). Parasite eggs were assigned to morphotypes defined by size and morphological characters, and the family was determined if possible from the egg morphology. To quantify the number of eggs we applied a modification of the McMaster flotation technique (Gordon and Whitlock 1939), which uses a very dense potassium iodide solution to enhance the ability to detect heavy weighed eggs (Meyer-Lucht and Sommer 2005). We recorded the number of different helminth infections in the whole population (parasite diversity) as well as helminth prevalence (percentage of infected individuals/examined individuals). On the individual level, we measured the number of different helminth infections per individual (NHI), the helminth prevalence (being infected or not) and the intensity of nematode infection (nematode eggs/g faeces). Due to their extremely low prevalence trematodes were not considered. In cestodes, numerical variation in egg release can be extremely high; therefore intensity of cestode infection was not included.
Software and data analyses
Mega 4.0 (Tamura et al. 2007) was used to edit nucleotide sequences manually, to align them, and to calculate amino acid differences between the alleles. As the number of amino acid differences between MHC alleles might not be a good predictor for their functional differences, we grouped MHC alleles according to similar antigen-binding motifs into so called MHC supertypes (Supplement Fig. 1). The technique was invented in human vaccine development studies and is supported by the fact that in humans different HLA alleles overlap in their peptide binding specifities (Southwood et al. 1998; Sette and Sidney 1999; Trachtenberg et al. 2003). This technique requires a minimum number of alleles, which was not observed in M. incanus (see below). For G. microtarsus, we followed a procedure proposed by Doytchinova and Flower (2005), which was successfully applied in a study on a natural primate population by Schwensow et al. (2007). In a first step amino acid sites under positive selection were identified, which are presumably involved in the antigen binding process, using the program CodeML implemented in PAML 3.15 (Yang 1997). For details on the calculations see Meyer-Lucht et al. (2008). In a second step, for each Grmi-DAB allele these positively selected amino acid sites were characterized by five z-descriptors (Sandberg et al. 1998) based on their lipophilic, steric and electronic properties: z1 (hydrophobicity), z2 (steric bulk), z3 (polarity), z4 and z5 (electronic effects). For each site these five z-values were assigned and a hierarchical cluster was calculated using the Euclidian distance method. The resulting groups (= MHC supertypes) possess similar physicochemical binding features at the putative antigen binding sites. These MHC supertypes were included as predictors in subsequent analyses.
The individuals were trapped in six forest sites of different size. We tested if there was an effect of the forest patch size on immune gene diversity to account for area effects. Therefore, we calculated linear regression analyses of the number of MHC alleles found per forest site in relation to the forest patch size. To compare the parasite loads between the two host species, we based our calculations on the total sample sizes of 102 G. microtarsus and 123 M. incanus individuals. The number of different helminth infections between the two host species was analysed by the non parametric Mann–Whitney U test. The helminth prevalence in the two hosts was compared by a χ2-test. The values for nematode infection intensity were log-transformed to account for heterogeneity, and comparison between species was again carried out with the Mann–Whitney U test.
To understand selection processes acting on the marsupial MHC DAB under natural conditions we investigated the relationship between the individual MHC DAB constitution and the parasite load in G. microtarsus and M. incanus. Therefore we used generalized linear models (GLMs) and included environmental, biotic and genetic predictors to explain different measures of parasite burden. As environmental predictors the categorical variables ‘forest site’ and ‘capture session’ were included, biotic predictors comprised the individual body condition (body mass index, BMI) and the sex. BMI was obtained from regressing body mass on body size (length of tibia), and using the residuals from this regression (Schulte-Hostedde et al. 2005). For G. microtarsus we used the presence/absence of 13 MHC DAB supertypes that occurred in frequencies >0.05 and <0.95 as genetic predictors. In addition we used the number of different MHC supertypes per individual as a measure for individual MHC diversity. In the case of M. incanus we used the presence/absence of the detected MHC DAB alleles, with Main-DAB*03 and Main-DAB*04 combined to one predictor (see above), and a combination of Main-DAB*07 and Main-DAB*08 because they always co-occurred. As a measure for individual MHC diversity the number of different MHC DAB alleles per individual was used. To avoid collinearity we tested all predictors for correlations and, if necessary, included them in separate, otherwise identical models. Missing data in the predictors BMI or sex reduced the number of included cases to 96 in G. microtarsus and 121 in M. incanus.
We fitted models for the number of different helminth infections per individual (NHI), the helminth prevalence and the nematode infection intensity (FEC). The models for NHI were calculated with a poisson error distribution and log link function. For prevalence data, logistic regression models were applied with a binomial error distribution and logit link function. For the infection intensity data, we used a quasipoisson error distribution with a log link function, which accounts for overdispersion in these data. We started with the full model including all predictor variables and conducted backward selection to find the minimal adequate model to explain our data. Backward selection was performed by dropping sequentially non significant predictors from the model. The new, less complex model was compared with the previous, more complex model by testing the change in deviance for significance. If the simplification was not associated to a significant increase in deviance, the less complex model was preferred. When all predictors had to be excluded, no model was better in explaining the data than the null model, i.e. there were no effects of any predictor on the response variable. The adjusted R2 was calculated as 1 − (residual deviance/residual df)/(null model deviance/null model df). Odds ratio estimates were calculated by exponentiation of the coefficient from the logistic regression model. Statistical tests were performed in R (version 2.7.0) (R Development Core Team 2008) and SPSS 16.0, using a threshold of P = 0.05.
Results
MHC DAB diversity
Gracilinanus microtarsus
The 102 individuals of G. microtarsus revealed high allelic diversity at the MHC DAB, with a total of 80 Grmi-DAB alleles (Table 1). We have described 47 of these alleles in an earlier study on a smaller sample size (N = 54, Meyer-Lucht et al. 2008, EU350150–EU350196). The 33 additional Grmi-DAB sequences were deposited at GenBank under FJ374838–FJ374870. Five groups of nucleotide sequences translated each into one identical amino acid sequence (Grmi-DAB*01a, *01b, *01c & *01d; Grmi-DAB*10a, *10b, *10c & *10d; Grmi-DAB*12a & *12b; Grmi-DAB*19a & *19b and Grmi-DAB*40a & *40b). Three alleles featured stop codons (Grmi-DAB*01e, *04 and *17) and two alleles (Grmi-DAB*37 and *49) carried a deletion of two nucleotides leading to a frame shift. These five alleles were regarded as potential pseudogenes and excluded from further calculations. In the remaining 75 Grmi-DAB alleles, sequence divergence in terms of variable amino acid sites and number of amino acid differences between alleles was high (Table 1). The number of alleles per individual ranged from one to nine, confirming our previous conclusion of multiple duplicated DAB loci in this species and identifying at least five MHC DAB loci in G. microtarsus. The number of different Grmi-DAB alleles on the amino acid level varied per forest site from 21 to 38 (Table 2). We did not detect an effect of the forest patch size on the number of MHC alleles in a forest site (R2 = 0.466, N = 5, ANOVA, F = 2.623, n.s.).
For further analyses of the relationship between the individual MHC DAB constitution and the parasite load we clustered the 75 Grmi-DAB alleles into 17 functional MHC supertypes based on similar antigen-binding motifs (Supplement Fig. 1) (Doytchinova and Flower 2005). Supertypes ST11, ST16 and ST17 were not included in subsequent calculation because they occurred in frequencies <0.05. ST01 was likewise excluded because it was present in all but three individuals (>0.95) and therefore not informative.
Marmosops incanus
In contrast to the diversity in G. microtarsus, only eight different Main-DAB alleles were detected in the total sample of 123 M. incanus individuals (EU350142–EU350149, Table 1). All of them were already known from our earlier study (N = 56, Meyer-Lucht et al. 2008). This means, extending the sample size to more than double did not reveal a single new allele. However, average sequence divergence among these eight Main-DAB alleles was similar to the divergence in the alleles of G. microtarsus (Table 1). Within a single individual two to four Main-DAB alleles were identified. In the six forest sites the number of Main-DAB alleles varied from five to seven (Table 2). There was no significant effect of the size of the forest site on the number of MHC alleles per forest site (R2 = 0.033, N = 6, ANOVA, F = 0.137, n.s.).
Parasite load
Gracilinanus microtarsus
In 102 faecal samples of G. microtarsus, we detected eleven distinct helminth egg morphotypes. Eight of these were classified as nematodes, among them six morphotypes belonging to the group of strongyle nematodes. The remaining two nematode morphotypes could not be further assigned. In addition, two different cestode morphotypes from the family Hymenolepidae were detected. In two individuals of G. microtarsus one trematode morphotype was detected with a single egg. This morphotype was excluded from the analyses due to its very low frequency.
Individuals carried zero (26.4%), one (49.0%), two (16.7%), three (6.9%) or four (1.0%) helminth infections. The most common parasite morphotype was the strongyle nematode N1, with a prevalence of 38.2% (Fig. 1a), followed in frequency by the nematode N3 (30.4%) and the cestode C1 (22.5%). The remaining morphotypes were rarely represented. They were added to the group ‘others’ unless they were shared with M. incanus.
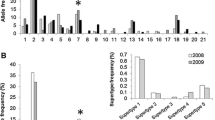
Parasite load with gastrointestinal nematodes and cestodes in G. microtarsus (N = 102, black bars) and M. incanus (N = 123, hatched bars). ‘Others’ represent nematode morphotypes that occurred in low frequencies and were not shared between host species. a Prevalence, b Intensity of infection (only nematodes). Bars illustrate the mean log-transformed FEC values [log(nematode eggs +1)] and the standard deviation
Marmosops incanus
In 123 M. incanus individuals the faecal samples revealed thirteen distinct helminth egg morphotypes, thus parasite diversity in the species was slightly higher than in G. microtarsus. Eleven of these were nematodes, among them five morphotypes of strongyle nematodes, one of the family Trichuridae (Capillaria spec.) and one of the family Oxyuridae (Syphacia spec.). The remaining four nematode morphotypes could not be assigned to the family level. Moreover, the same two cestode morphotypes were identified as found in G. microtarsus (Fig. 1a).
M. incanus individuals carried one (63.4%), two (31.7%) or three (4.9%) helminth infections. The strongyle nematode N1 was prevalent in all M. incanus individuals (Fig. 1a). The second most prevalent parasite was the cestode C1 (13.8%) followed by the nematode N2 (10.6%). The remaining morphotypes occurred sporadically in one to three samples and were merged to ‘others’, if they were not shared between host species. N1 reached very high infection intensities in M. incanus, whereas the other morphotypes occurred in low quantities (Fig. 1b).
The two host species shared six helminth morphotypes, four nematodes (N1, N2, N4, N5) and the two cestodes C1 and C2 (Fig. 1a). The differences in individual parasite load between the two marsupial species were highly significant: the number of different helminth infections (NHI) in M. incanus individuals was higher than in G. microtarsus individuals (Mann–Whitney U test; Z = −3.92; P < 0.001). The helminth prevalence (χ2 = 55.1, df = 1, P < 0.001) and nematode infection intensity (Mann–Whitney U test; Z = −9.03; P < 0.001) in M. incanus exceeded by far the corresponding measures in G. microtarsus, too (Fig. 1a, b).
Factors influencing the parasite load
Gracilinanus microtarsus
In terms of environmental predictors, the capture session explained a substantial part of the variation regarding both the number of different helminth infections and the nematode infection intensity. In these models capture session 5 was associated with increased parasitism, as removing this term from the model led to a significant increase in deviation (NHI: z = 2.59, df = 90, P = 0.009; nematode infection intensity: t = 2.51, df = 91, P = 0.014; Table 3). Neither the BMI nor the sex of the host had a significant influence on the different measures of parasitism in G. microtarsus.
Regarding genetic predictors, the number of different helminth infections was influenced by the presence or absence of ST09. The supertype was associated to a lower number of different nematode infections (z = −2.07, df = 90, P = 0.039). In terms of helminth prevalence two MHC supertypes were identified to be influential. The chance of being helminth infected was reduced to an odds ratio (OR) of 0.24 in presence of ST09 (z = −2.20, df = 93, P = 0.028). In the same way, ST14 reduced the helminth infection probability (z = −2.09, df = 93, P = 0.037; OR = 0.18).
Marmosops incanus
No predictor revealed a significant effect on the number of different helminth infections, as no model explained the variance in the data better than the null model. No model could be calculated for helminth prevalence because all individuals were infected. The nematode infection intensity varied between sites in M. incanus; site 3 and the control site were associated to increased infection intensities (t = 2.08, df = 114, P = 0.040 and t = 5.72, df = 114, P < 0.001, respectively, Table 3). Again, neither host BMI nor sex had a significant influence on parasitism. In terms of genetic predictors, the presence of allele Main-DAB*05 increased the probability of high nematode infection intensities (t = 2.90, df = 114, P = 0.005).
Discussion
In this study we investigated possible causes and consequences of low MHC diversity by analysing gastrointestinal parasite burden and functional associations between the individual MHC constitution and parasite load in two species of mouse opossums. We tested two scenarios under which low MHC diversity could occur. We assumed that the species with low MHC diversity could be under relaxed selection pressure to maintain immune gene diversity and predicted low parasite diversity (‘Evolutionary equilibrium’ scenario). In an alternative scenario (‘Unbalanced situation’ scenario), we assumed that a recent loss in MHC diversity lead to a lack of resistance alleles and predicted an increased parasite burden in the species with low MHC diversity. We found that parasite diversity (the total number of helminth morphotypes) between the two host species was not markedly different. However, considerable differences in the individual parasite loads (parasite prevalence and infection intensity) between the two species were observed: while M. incanus revealed low MHC DAB diversity and high parasite load, G. microtarsus showed a tenfold higher population wide MHC DAB diversity and lower parasite burden supporting the second scenario.
MHC DAB diversity in Gracilinanus microtarsus and Marmosops incanus
One of the most prominent features of the MHC is the enormous variability found in the majority of vertebrates studied to date (Klein 1986; Hedrick 1994; Apanius et al. 1997; Sommer 2005; Piertney and Oliver 2006). The extensive MHC DAB diversity detected in G. microtarsus resembles high levels of MHC II diversity that are usually observed in wild populations of eutherians (summarized in O’Brien and Yuhki 1999; Bernatchez and Landry 2003; Sommer 2005; Piertney and Oliver 2006). Moreover, the high number of at least five DAB loci in G. microtarsus suggests the presence of a mechanism for generating MHC diversity that is known, for example, from the rhesus macaque (Macaca mulatta) and the Californian sea lion (Zalophus californianus). In contrast to high levels of allelic variation from a single MHC gene locus, this organisation comprises multiple duplicated MHC loci, each of them with limited variability, and present in variable configurations between individuals (Doxiadis et al. 2000; Bowen et al. 2004). However, for marsupials very limited polymorphism at the MHC class II was assumed to be characteristic, inferred from marsupials used in laboratory investigations (Stone et al. 1996, 1998). Most molecular studies on marsupial MHC indeed revealed low class II variation (using RFLP: McKenzie and Cooper 1994; using cDNA library: Schneider et al. 1991; Stone et al. 1999; Lam et al. 2001; Siddle et al. 2007b). G. microtarsus is the first marsupial that shows this explicit MHC class II polymorphism and signs of positive selection (Meyer-Lucht et al. 2008). But it does not appear to be a South American exception, as Holland et al. (2008a, b) recently described considerable MHC class II variation in an Australian marsupial species: the brushtail possum (T. vulpecula).
In contrast, limited MHC DAB polymorphism was indeed found in our second study species, M. incanus, which is concordant with the supposed general low MHC class II diversity in marsupials. One might argue that the low variation in M. incanus might be of methodological origin: the existence of null alleles. Although the occurrence of null alleles in both host species cannot be excluded, we do not assume a major impact on our results for the reasons discussed elsewhere (Meyer-Lucht et al. 2008).
In general, interpretations of low MHC polymorphism in natural populations refer to constraints of the mating system (Sommer 2003), reduced selection pressure by pathogens (Slade and McCallum 1992) or loss of genetic diversity through inbreeding or genetic drift in restricted populations (Klein 1986; Sommer 2005). While we can exclude constraints of different mating systems in M. incanus and G. microtarsus because both species share the same partially semelparous mating behaviour (Lorini et al. 1994; Martins et al. 2006), we addressed the explanations of a lowered selection pressure and lost genetic diversity by investigations of the parasite load.
Parasite load in Gracilinanus microtarsus and Marmosops incanus
On the population level, parasite diversity between the host species was not conspicuously different as we detected thirteen helminth morphotypes in M. incanus and eleven in G. microtarsus. Moreover, the two host species showed a similar fauna of gastrointestinal helminthes: six parasite morphotypes were shared, among them two frequent ones. Individual parasite prevalence and infection intensity was higher in M. incanus, the species featuring low immune gene variation.
Parasitism is a highly complex and dynamic process dependent on a multitude of factors. The differences in the parasite load between the two species may be affected by their different microhabitat preferences. While G. microtarsus is mainly arboreal and uses the canopy, the terrestrial locomotion of M. incanus might increase its infection risk due to a greater exposure to soil borne parasites or those with faecal-oral transmission (Nunn et al. 2003). It is well known that spatial and temporal environmental variation plays an important role in parasite prevalence and intensity, beside the immune state, sex, and age of the host (Abu-Madi et al. 2000; Behnke et al. 2001). In our study, the statistical models indicated that environmental factors, such as the forest sites or the capture sessions, influence the parasite load in both species. However, we did not detect a strong influence of the BMI or the sex on parasite load in either species, although there is a well documented association between testosterone and the immune system (summarized in Zuk and McKean 1996). Differences in parasite load between sexes are known from a number of studies on vertebrates. Sexually mature male vertebrates are often more susceptible to infection and carry higher parasite burdens, while estrogens even stimulate immunity (reviewed in Poulin 1996; Zuk and McKean 1996). But such sex biased differences in parasite loads might be small and therefore very difficult to detect (Zuk and McKean 1996).
Parasite load and functional associations with host genetics
In both species, our data provide evidence that the parasite load is affected by host genetics. We confirmed functional associations between specific MHC variants and the individual parasite load. Beneficial MHC variants were detected in G. microtarsus: the presence of ST09 and ST14 was correlated to reduced parasite infections. In ST09 this association was due to a single allele (Grmi-DAB*16), whereas in ST14 not a single allele, but the whole group accounted for the positive effect (Supplement Fig. 1). On the other hand, the allele Main-DAB*05 in M. incanus appeared to be disadvantageous, as it was identified to increase parasite infection intensity. We do not have information on the expression of the MHC DAB alleles, as RNA samples are not available. However, the genomic level shows a comprehensive pattern of all available MHC variants, independent of temporal variation in expression. The existence of beneficial and disadvantageous MHC variants is explained by dynamic processes between host and parasites. Natural selection will favour pathogens that are not recognized by the most common MHC molecules, so there is selection for rare or new MHC alleles and against common ones in the host population (May and Anderson 1990; Takahata and Nei 1990). These functional associations between MHC variants and high or low parasite loads mirror the classical pattern of pathogen-driven selection acting on the MHC. Beneficial and/or disadvantageous MHC alleles with regard to parasite infections were detected in several studies (e.g. Schwaiger et al. 1995; Langefors et al. 2001; Lohm et al. 2002; Froeschke and Sommer 2005; Harf and Sommer 2005; Meyer-Lucht and Sommer 2005; Schad et al. 2005; Westerdahl et al. 2005). Recent studies also demonstrated associations between certain MHC supertypes and diseases resistance or susceptibility (Trachtenberg et al. 2003; Schwensow et al. 2007). Here, we provide the first study indicating that pathogen-driven selection acts on the MHC class II of natural marsupial populations.
We did not find indication for a larger individual number of different MHC variants being advantageous in terms of parasite defence, which would correspond in a broader sense to the hypothesis of heterozygote advantage (Doherty and Zinkernagel 1975). According to this hypothesis, heterozygous individuals are in selective advantage over homozygous ones, because they are able to detect a broader range of parasites as a result of a larger number of different MHC molecules and thereby obtain a higher parasite resistance.
Low MHC diversity: evolutionary equilibrium or unbalanced situation?
Parasite diversity in the species with low MHC diversity, M. incanus, was not lower but even higher than in G. microtarsus. We therefore reject the first scenario predicting a relaxed pathogenic selection pressure accounting for the low MHC diversity in M. incanus. On the contrary, this species was distinctly higher parasitized. In the state of evolutionary equilibrium, host species exposed to a diverse array of parasites should harbour a variety of resistance alleles or a repertoire of inducible defences (Altizer et al. 2003). With its numerous MHC DAB alleles G. microtarsus seems to be well equipped, but this is not fulfilled in M. incanus.
Parasite load in M. incanus seems to be rather the consequence than the cause for its low MHC diversity. The explicitly higher helminth prevalence and infection intensities in M. incanus could be a result of its genetic homogeneity and low genotypic variation because genetic uniformity facilitates the spreading of pathogens through a population (Meagher 1999). The pattern of a relative low MHC II diversity combined with a relative high parasite load in M. incanus might be a sign of an unbalanced situation, i.e. a recent loss of genetic diversity. Thereby, M. incanus could have lost resistance alleles or other important parts of its adaptive evolutionary potential.
Some species are known to perform very well despite low MHC variation or even monomorphism and show no signs of severe infectious diseases, e.g. moose (Alces alces) or mountain goats (Oreamnos americanus) (Mikko and Anderson 1995; Mainguy et al. 2007). However, the pattern of low MHC class II variation and high parasite load in M. incanus resembles findings from other prominent species, for instance, the giant panda (Ailuropoda melanoleuca), which is particularly susceptible to infectious diseases and parasites. Wan et al. (2006) found only seven MHC DRB alleles in 60 individuals, and credited this relatively low variation to a genetic bottleneck. Moreover, a recent study on the Tasmanian devil (S. harrisii) detected a severely depleted MHC class I and ascribed the easy spreading of a contagious tumour (devil facial tumour disease) to this depletion, which is currently putting the Tasmanian devil under threat of extinction (Siddle et al. 2007a, but see Murchison et al. 2010).
Other examples of mammals with extremely low MHC variation due to genetic bottlenecks are, for instance, cheetahs (Acinonyx jubatus, O’Brien et al. 1985), Asian lions (Panthera leo persica, Yuhki and O’Brien 1990) or Przewalski’s horses (Equus przewalskii, Hedrick et al. 1999). In these cases, it is assumed that genetic drift was stronger than balancing selection in shaping MHC variation (Sommer 2005). As these species revealed very low levels of neutral genetic variation, too, it cannot be discriminated between effects based on reduced MHC variation and effects attributed to low variation at other fitness relevant loci (O’Brien et al. 1985; Yuhki and O’Brien 1990; Hedrick et al. 1999).
We have no evidence for a population bottleneck in M. incanus. Ecological studies, however, indicated that M. incanus may be more sensitive to habitat fragmentation than G. microtarsus, since M. incanus is restricted to patches of native vegetation (Umetsu and Pardini 2007) and prefers old, undisturbed forest with dense canopy cover (Püttker et al. 2008). In small and isolated fragments M. incanus decreases in abundance (Pardini et al. 2005). G. microtarsus, on the other hand, is not less abundant in small or isolated fragments (Pardini et al. 2005), prefers vegetation of disturbed forest (Püttker et al. 2008) and has even been captured in eucalyptus plantations (Umetsu and Pardini 2007). Ecological constraints might be responsible for a reduced migration between forest fragments in M. incanus, leading to lower gene flow and loss of genetic variation as a consequence of genetic drift. However, as only very few alleles were also detected in the control site and no effect of the forest patch size on the number of MHC alleles was visible, fragmentation sensitivity is not sufficient to explain the observed low variation in M. incanus.
To further investigate whether a bottleneck event and/or phylogenetic constraints account for the low MHC II variability in M. incanus, a deeper survey of the genetic diversity in this species is required, including neutral markers such as microsatellites, as well as more studies on marsupial species. In future studies we aim to compare M. incanus populations from small fragments in different landscapes surrounded by 25–35%, 45–55% and 85–100% of forest habitat to discover whether the low level of immune gene variation in M. incanus is characteristic for this species or result of a local bottleneck event.
In conclusion, our study revealed a relatively low parasite load in the species with high MHC DAB diversity, whereas very high parasite burdens were detected in the opossum showing low immune gene diversity. In both species it became apparent that the MHC class II is functionally important in the individual disease defence against gastrointestinal helminths, which has been shown here for the first time in marsupials. The question whether or not low MHC class II diversity is a phylogenetic characteristic of marsupials will not be resolved until substantial knowledge on the marsupial MHC in natural populations is gathered. It may be that the high diversity in G. microtarsus and not the low level of diversity in M. incanus is an exception to the rule. However, a definite explanation for the remarkable differences in MHC diversity in marsupials needs further investigations. Research on the marsupial MHC has only just begun.
References
Abu-Madi MA, Behnke JM, Lewis JW, Gilbert FS (2000) Seasonal and site specific variation in the component community structure of intestinal helminths in Apodemus sylvaticus from three contrasting habitats in south-east England. J Helminthol 74:7–15
Altizer S, Harvell D, Friedle E (2003) Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol Evol 18:589–596
Altizer S, Nunn CL, Lindenfors P (2007) Do threatened hosts have fewer parasites? A comparative study in primates. J Anim Ecol 76:304–314
Apanius V, Penn D, Slev P, Ruff L, Potts W (1997) The nature of selection on the major histocompatibility complex. Crit Rev Immunol 17:179–224
Behnke JM, Barnard CJ, Bajer A, Bray D, Dinmore J, Frake K, Osmond J, Race T, Sinski E (2001) Variation in the helminth community structure in bank voles (Clethrionomys glareolus) from three comparable localities in the Mazury Lake District region of Poland. Parasitology 123:401–414
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16:363–377
Bowen L, Aldridge BM, Gulland F, Van Bonn W, DeLong R, Melin S, Lowenstine LJ, Stott JL, Johnson ML (2004) Class II multiformity generated by variable MHC-DRB region configurations in the California sea lion (Zalophus californianus). Immunogenetics 56:12–27
Brown JL, Erklund A (1994) Kin recognition and the major histocompatibility complex: an integrative review. Am Nat 143:435–461
Brown JH, Jardetzky TS, Gorga JC, Stern LJ (1993) Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364:33–39
Caceres NC (2004) Diet of three didelphid marsupials (Mammalia, Didelphimorphia) in southern Brazil. Mamm Biol 69:430–433
Cassinello J, Gomendio M, Roldan E (2001) Relationship between coefficient of inbreeding and parasite burden in endangered gazelles. Conserv Biol 15:1171–1174
Coltman D, Pilkington J, Smith J, Pemberton J (1999) Parasite-mediated selection against inbred soay sheep in a free-living, island population. Evolution 53:1259–1267
Cunha AA, Vieira MV (2002) Support diameter, incline, and vertical movements of four didelphid marsupials in the Atlantic forest of Brazil. J Zool 258:419–426
Doherty PC, Zinkernagel RM (1975) Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256:50–52
Dorman SE, Hatem CL, Tyagi S, Aird K, Lopez-Molina J, Pitt MLM, Zook BC, Dannenberg AM Jr., Bishai WR, Manabe YC (2004) Susceptibility to tuberculosis: clues from studies with inbred and outbred New Zealand white rabbits. Infect Immun 72:1700–1705
Doxiadis GGM, Otting N, de Groot NG, Noort R, Bontrop RE (2000) Unprecedented polymorphism of MHC-DRB region configurations in Rhesus Macaques. J Immunol 164:3193–3199
Doytchinova IA, Flower DR (2005) In silico identification of supertypes for class II MHCs. J Immunol 174:7085–7095
Ferrari N, Cattadori I, Nespereira J, Rizzoli A, Hudson P (2004) The role of host sex in parasite dynamics: field experiments on the yellow-necked mouse Apodemus flavicollis. Ecol Lett 7:88–94
Fonseca GAB, Kierulff MCM (1989) Biology and natural history of Brazilian Atlantic forest small mammals. Bull Fla State Mus Biol Sci 34:99–152
Froeschke G, Sommer S (2005) MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the southern Kalahari. Mol Biol Evol 22:1254–1259
Gordon HM, Whitlock HV (1939) A new technique for counting nematode eggs in sheep feaces. J Counc Sci Ind Res Melbourne 12:50–52
Goüy de Bellocq J, Charbonnel N, Morand S (2008) Coevolutionary relationship between helminth diversity and MHC class II polymorphism in rodents. J Evol Biol 21:1144–1150
Harf R, Sommer S (2005) Association between MHC class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the southern Kalahari. Mol Ecol 14:85–91
Hedrick PW (1994) Evolutionary genetics of the major histocompatibility complex. Am Nat 143:945–964
Hedrick PW (2002) Pathogen resistance and genetic variation at MHC loci. Evolution 56:1902–1908
Hedrick PW, Parker KM, Miller EL, Miller PS (1999) Major histocompatibility complex variation in the endangered Przewalski’s Horse. Genetics 152:1701–1710
Holland O, Cowan P, Gleeson D, Chamley L (2008a) High variability in the MHC class II DA beta chain of the brushtail possum (Trichosurus vulpecula). Immunogenetics 60:775–781
Holland O, Cowan P, Gleeson D, Chamley L (2008b) Novel alleles in classical major histocompatibility complex class II loci of the brushtail possum (Trichosurus vulpecula). Immunogenetics 60:449–460
Hughes AL, Yeager M (1998) Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet 32:415–434
Klein J (1986) Natural history of the major histocompatibility complex. Wiley & Sons, New York
Klein J, Horejsi V (1997) Immunology. Blackwell Science, Oxford
Kurtz J, Kalbe M, Aeschlimann PB, Häberli MA, Wegner KM, Reusch TBH, Milinski M (2004) Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proc R Soc Lond B Biol Sci 271:197–204
Lam MK-P, Belova K, Harrison GA, Coopera D (2001) Cloning of the MHC class II DRB cDNA from the brushtail possum (Trichosurus vulpecula). Immunol Lett 76:31–36
Langefors Å, Lohm J, Grahn M, Andersen Ø, von Schantz T (2001) Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonica in Atlantic salmon. Proc R Soc Lond B Biol Sci 268:479–485
Liersch S, Schmid-Hempel P (1998) Genetic variation within social insect colonies reduces parasite load. P Roy Soc Lond B Biol Sci 265:221-225
Lohm J, Grahn M, Langefors Å, Anderson Ø, Storset A, von Schantz T (2002) Experimental evidence for major histocompatibility complex-allele-specific resistance to a bacterial infection. Proc R Soc Lond B Biol Sci 269:2029–2033
Lorini ML, Oliveira JA, Persson VG (1994) Annual age structure and reproductive patterns in Marmosa incana (Lund, 1841) (Didelphidae, Marsupialia). Mamm Biol 59:65–73
Mainguy J, Worley K, Côté S, Coltman D (2007) Low MHC DRB class II diversity in the mountain goat: past bottlenecks and possible role of pathogens and parasites. Conserv Genet 8:885–891
Martins EG, Bonato V (2004) On the diet of Gracilinanus microtarsus (Marsupialia, Didelphidae) in an Atlantic Rainforest fragment in south-eastern Brazil. Mamm Biol 69:58–60
Martins EG, Bonato V, da Silva CQ, dos Reis SF (2006) Partial semelparity in the neotropical didelphid marsupial Gracilinanus microtarsus. J Mammal 87:915–920
McCallum H, Dobson A (1995) Detecting disease and parasite threats to endangered species and ecosystems. Trends Ecol Evol 10:190–194
McKenzie LM, Cooper DW (1994) Low MHC class II variability in a marsupial. Reprod Fertil Dev 6:721–726
Meagher S (1999) Genetic diversity and Capillaria hepatica (Nematoda) prevalence in Michigan deer mouse populations. Evolution 53:1318–1324
Meyer-Lucht Y, Sommer S (2005) MHC diversity and the association to nematode parasitism in the yellow-necked mouse (Apodemus flavicollis). Mol Ecol 14:2233–2243
Meyer-Lucht Y, Otten C, Püttker T, Sommer S (2008) Selection, diversity and evolutionary patterns of the MHC class II DAB in free-ranging Neotropical marsupials. BMC Genet 9:39
Mikko S, Anderson L (1995) Low major histocompatibility complex class II diversity in European and North American moose. Proc Natl Acad Sci USA 92:4259–4263
Murchison EP, Tovar C, Hsu A et al (2010) The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science 327:84–87
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Nunn CL, Altizer S, Jones KE, Sechrest W (2003) Comparative tests of parasite species richness in primates. Am Nat 162:597–614
O’Brien SJ, Yuhki N (1999) Comparative genome organization of the major histocompatibility complex: lessons from the Felidae. Immunol Rev 167:133–144
O’Brien SJ, Roelke ME, Marker L, Newman A, Winkler CA, Meltzer D, Colly L, Evermann JF, Bush M, Wildt DE (1985) Genetic basis for species vulnerability in the cheetah. Science 227:1428–1434
Pardini R, Marques de Souza S, Braga-Neto R, Metzger JP (2005) The role of forest structure, fragment size and corridors in maintaining small mammal abundance and diversity in an Atlantic forest landscape. Biol Conserv 124:253–266
Piertney SB, Oliver MK (2006) The evolutionary ecology of the major histocompatibility complex. Heredity 96:7–21
Poulin R (1996) Sexual inequalities in helminth infections: a cost of being a male? Am Nat 147:287
Prugnolle F, Manica A, Charpentier M, Guégan JF, Guernier V, Balloux F (2005) Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol 15:1022–1027
Püttker T, Meyer-Lucht Y, Sommer S (2006) Movement distances of five rodent and two marsupial species in forest fragments of the coastal Atlantic rainforest, Brazil. Ecotropica 12:131–139
Püttker T, Pardini R, Meyer-Lucht Y, Sommer S (2008) Responses of five small mammal species to micro-scale variations in vegetation structure in secondary Atlantic forest remnants, Brazil. BMC Ecol 8:9
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni F, Hirota M (2009) The Brazilian Atlantic forest: how much is left, and how is the remaining forest distributed? Implications for conservation. Biol Conserv 142:1141–1153
Sandberg M, Eriksson L, Jonsson J, Sjostrom M, Wold S (1998) New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J Med Chem 41:2481–2491
Schad J, Sommer S, Ganzhorn J (2004) MHC variability of a small lemur in the littoral forest fragments of south-eastern Madagascar. Conserv Genet 5:299–309
Schad J, Ganzhorn JU, Sommer S (2005) MHC constitution and parasite burden in the Malagasy mouse lemur, Microcebus murinus. Evolution 59:439–450
Schneider S, Vincek V, Tichy H, Figueroa F, Klein J (1991) MHC class II genes of a marsupial, the red-necked wallaby (Macropus rufogriseus): identification of new gene families. Mol Biol Evol 8:753–766
Schulte-Hostedde AI, Zinner B, Millar JS, Hickling GJ (2005) Restitution of mass-size residuals: validating body condition indices. Ecology 86:155–163
Schwaiger FW, Gostomski D, Stear MJ (1995) An ovine major histocompatibility complex DRB1 allele is associated with low fecal counts following natural, predominantly Ostertagia circumcincta infection. Int J Parasitol 25:815–822
Schwensow N, Fietz J, Dausmann K, Sommer S (2007) Neutral versus adaptive genetic variation in parasite resistance: importance of MHC-supertypes in a free-ranging primate. Heredity 99:265–277
Seivwright LJ, Redpath SM, Mougeot F, Watt L, Hudson PJ (2004) Faecal egg counts provide a reliable measure of Trichostrongylus tenuis intensities in free-living red grouse Lagopus lagopus scoticus. J Helminthol 78:69–76
Sette A, Sidney J (1999) Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201–212
Siddle H, Kreiss A, Eldridge MDB, Noonan E, Clarke CJ, Pyecroft S, Woods GM, Belov K (2007a) From the cover: transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc Natl Acad Sci USA 104:16221–16226
Siddle H, Sanderson C, Belov K (2007b) Characterization of major histocompatibility complex class I and class II genes from the Tasmanian devil (Sarcophilus harrisii). Immunogenetics 59:753–760
Slade RW, McCallum HI (1992) Overdominant vs frequency-dependent selection at MHC loci. Genetics 132:861–862
Sommer S (2003) Effects of habitat fragmentation and changes of dispersal behaviour after a recent population decline on the genetic variability of noncoding and coding DNA of a monogamous Malagasy rodent. Mol Ecol 12:2845–2851
Sommer S (2005) The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool 2:16
SOS Mata Atlântica and Instituto Nacional de Pesquisas Espaciais (2008) Atlas dos remanescentes florestais da Mata Atlântica, período de 2000 a 2005
Soulsby EJL (1982) Helminths, arthropods and protozoa of domesticated animals, Lea & Febiger, Philadelphia
Southwood S, Sidney J, Kondo A, del Guercio M-F, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A (1998) Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 160:3363–3373
Spielman D, Brook B, Briscoe D, Frankham R (2004) Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet 5:439–448
Stear MJ, Bishop SC, Doligalska M, Duncan JL, Holmes PH, Irvine J, McCririe L, McKellar QA, Sinski E, Murray M (1995) Regulation of egg production, worm burden, worm length and worm fecundity by host responses in sheep infected with Ostertagia circumcincta. Parasite Immunol 17:643–652
Stear MJ, Bairden K, Duncan JL, Holmes PH, McKellar QA, Park M, Strain S, Murray M, Bishop SC, Gettinby G (1997) How hosts control worms. Nature 389:27
Stone W, Bruun D, Manis G, Holste S, Hoffman E, Spong K, Walunas T (1996) The immunobiology of the marsupial, Monodelphis domestica. In: Stolen J, Fletcher T, Bayne C, Secombes C, Zelikoff J, Twerdok L, Anderson D (eds) Modulators of immune responses: the evolutionary trail. SOS Publications, Fair Haven, NJ, pp 149–165
Stone W, Brunn D, Foster E, Manis G, Hoffman E, Saphire D, VandeBerg J, Infante A (1998) Absence of a significant mixed lymphocyte reaction in a marsupial (Monodelphis domestica). Lab Anim Sci 48:184–189
Stone W, Bruun D, Fuqua C, Glass L, Reeves A, Holste S, Figueroa F (1999) Identification and sequence analysis of an Mhc class II B gene in a marsupial (Monodelphis domestica). Immunogenetics 49:461–463
Tabarelli M, Cardoso da Silva J, Gascon C (2004) Forest fragmentation, synergisms and the impoverishment of neotropical forests. Biodivers Conserv 13:1419–1425
Tabarelli M, Pinto LP, Silva JMC, Hirota M, Bede L (2005) Challenges and opportunities for biodiversity conservation in the Brazilian Atlantic forest. Conserv Biol 19:695–700
Takahata N, Nei M (1990) Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124:967–978
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Teixeira AMG, Soares-Filho BS, Freitas SR, Metzger JP (2009) Modeling landscape dynamics in an Atlantic Rainforest region: implications for conservation. Forest Ecol Manag 257:1219–1230
Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E, Funkhouser R, Fugate M, Theiler J, Hsu YS, Kunstman K, Wu S, Phair J, Erlich H, Wolinsky S (2003) Advantage of rare HLA supertype in HIV disease progression. Nat Med 9:928–935
Tregenza T, Wedell N (2000) Genetic compatibility, mate choice and patterns of parentage: invited review. Mol Ecol 9:1013–1027
Umetsu F, Pardini R (2007) Small mammals in a mosaic of forest remnants and anthropogenic habitats-evaluating matrix quality in an Atlantic forest landscape. Landsc Ecol 22:517–530
Vieira EM, Monteiro-Filho ELA (2003) Vertical stratification of small mammals in the Atlantic Rainforest of south-eastern Brazil. J Trop Ecol 19:501–507
Wan Q-H, Zhu L, Wu HUA, Fang S-G (2006) Major histocompatibility complex class II variation in the giant panda (Ailuropoda melanoleuca). Mol Ecol 15:2441–2450
Wegner KM, Kalbe M, Kurtz J, Reusch TBH, Milinski M (2003a) Parasite selection for immunogenetic optimality. Science 301:1343
Wegner KM, Reusch TBH, Kalbe M (2003b) Multiple parasites are driving major histocompatibility complex polymorphism in the wild. J Evol Biol 16:224–232
Westerdahl H, Waldenström J, Hansson B, Hasselquist D, von Schantz T, Bensch S (2005) Associations between malaria and MHC genes in a migratory songbird. Proc R Soc Lond B Biol Sci 272:1511–1518
Woodroffe R (1999) Managing disease threats to wild mammals. Anim Conserv 2:185–193
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yuhki N, O’Brien SJ (1990) DNA variation of the mammalian major histocompatibility complex reflects genomic diversity and population history. Proc Natl Acad Sci USA 87:836–840
Zuk M (1990) Reproductive strategies and disease susceptibility: an evolutionary viewpoint. Parasitol Today 6:231–233
Zuk M, McKean KA (1996) Sex differences in parasite infections: Patterns and processes. Int J Parasitol 26:1009–1024
Acknowledgements
Many thanks to Tamara Münkemüller for steady and patient advice in statistical questions. Further, we thank Fabiana Umetsu, Christoph Knogge, and Klaus Henle for scientific and logistic support. We are grateful to Birgit Bieber and Anke Schmidt for assistance in the genetic laboratory and to many field assistants for supporting field work. Financial support for this study was provided by the BMBF Germany within the BIOCAPSP project (German Federal Ministry of Education and Research, project ID: 01 LB 0202). This study is part of a cooperation supported by the Brazilian Council for Research and Technology (CNPq) (Project ‘Biodiversity conservation in fragmented landscapes on the Atlantic Plateau of São Paulo’, No. 590041/2006-1).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meyer-Lucht, Y., Otten, C., Püttker, T. et al. Variety matters: adaptive genetic diversity and parasite load in two mouse opossums from the Brazilian Atlantic forest. Conserv Genet 11, 2001–2013 (2010). https://doi.org/10.1007/s10592-010-0093-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0093-9