Abstract
Cutaneous malignant melanoma is the most lethal form of skin cancer, with 5-year survival rates of <5 % for patients presenting with metastatic disease. Mechanisms underlying metastatic spread of UVR-induced melanoma are not well understood, in part due to a paucity of animal models that accurately recapitulate the disease in its advanced forms. We have employed a transgenic mouse strain harboring a tandem deletion of the nm23-m1 and nm23-m2 genes to assess the combined contribution of these genes to suppression of melanoma metastasis. Crossing of the nm23-h1/nm23-h2 knockout in hemizygous-null form ([m1m2]+/−) to a transgenic mouse strain (hepatocyte growth factor/scatter factor-overexpressing, or HGF+ strain) vulnerable to poorly-metastatic, UVR-induced melanomas resulted in UVR-induced melanomas with high metastatic potential. Metastasis to draining lymph nodes was seen in almost all cases of back skin melanomas, while aggressive metastasis to lung, thoracic cavity, liver and bone also occurred. Interestingly, no differences were observed in the invasive characteristics of primary melanomas of HGF+ and HGF+ × [m1m2]+/− strains, with both exhibiting invasion into the dermis and subcutis, indicating factors other than simple invasive activity were responsible for metastasis of HGF+ × [m1m2]+/− melanomas. Stable cell lines were established from the primary and metastatic melanoma lesions from these mice, with HGF+ × [m1m2]+/− lines exhibiting increased single cell migration and genomic instability. These studies demonstrate for the first time in vivo a potent metastasis suppressor activity of NM23 in UVR-induced melanoma, and have provided new tools for identifying molecular mechanisms that underlie melanoma metastasis.
Similar content being viewed by others
Introduction
Cutaneous malignant melanoma (CMM) represents one of the most aggressive malignancies with its occurrence increasing worldwide at an alarming rate over the past 50 years [1]. Exposure to the ultraviolet radiation (UVR) spectrum in sunlight is well-established as a risk factor for melanoma, with intermittent sun exposure as a child and/or young adult associated with adult-onset malignancy [2, 3]. Currently, few treatment therapies are available for patients with metastatic melanoma, emphasizing the need for better understanding of the mechanisms underlying the disease in its advanced forms.
The first metastasis suppressor gene to be described was nm23-m1 (termed M1 isoform in mouse, H1 in human), initially identified by its diminished expression in metastatic variants of melanoma and breast carcinoma cell lines [4]. In these and a number of other human cancers, low NM23-H1 expression has been associated with increased tumor aggressiveness and poor clinical outcome [5–7]. While NM23 proteins participate in multiple cellular regulatory pathways, mechanism(s) underlying the metastasis suppressor activity of NM23-H1/M1 are not well-understood. The protein displays three distinct enzymatic activities in vitro, including its nucleoside diphosphate kinase (NDPK), histidine kinase and 3′–5′ exonuclease activities [8–10]. The histidine kinase and NDPK have both been suggested to participate in suppression of motile and invasive phenotypes in vitro, and these represent plausible targets of metastasis suppressor function. However, the NDPK and 3′–5′ exonuclease activities also suggest potential roles in DNA repair and/or replication, with NDPK possibly providing balance in nucleotide pools for DNA polymerase activity, and the 3′–5′ exonuclease functioning in proofreading during replication and/or repair. Along these lines, we demonstrated NM23 expression to be critical for genomic stability in the yeast Saccharomyces cerevisiae [11]. More recently, we observed important contributions of NM23-H1 and NM23-H2 to maintenance of genomic stability in human melanoma and mouse cell lines, both in basal and genotoxic conditions [12]. Importantly, mice rendered hemizygous-null at the tandemly-arranged nm23-m1 and nm23-m2 loci [m1m2]+/− exhibit vulnerability to UVR-induced melanoma in situ and epithelioid cyst formation on tail skin, providing in vivo evidence for the DNA repair function. These observations suggest that loss of nm23 expression contributes to both the genesis and progression of human melanoma.
To assess the role of NM23-M1 and NM23-M2 in melanoma progression in vivo, we have crossed [m1m2]+/− mice with a transgenic mouse strain engineered for overexpression of hepatocyte growth factor (HGF+). The HGF+ strain exhibits a form of UVR-induced melanoma with human characteristics, rapid growth but minimal metastatic potential [13]. The current study demonstrates that nm23-m1 and nm23-m2 deficiency confers high metastatic potential to melanomas of the HGF+ strain, with aggressively-growing metastases occurring at sites analogous to those seen in human melanoma (i.e. lymph node, lung, liver and bone). In addition, cell lines generated from melanomas of the HGF+ × [m1m2]+/− hybrid exhibit increased motility and genomic instability, implicating functional roles for NM23 proteins in melanoma progression.
Materials and methods
UVB/A radiation of mice and melanoma surveillance
Protocols for murine experiments were approved by the Institutional Animal Care and Use Committee at the University of Kentucky (Protocol 00801M2004). Mice were bred and genotyping was performed as described previously [12]. A C57BL/6-derived mouse strain (HGF+) overexpressing a melanocyte-targeted HGF transgene under control of the metallothionein promoter [13, 14] was utilized for these studies. Melanocytes in the HGF+ strain exhibit a “humanized” distribution of melanocytes within the epidermis, dermis, and epidermal-dermal junction, which renders them more susceptible to DNA damaging effects of UVR [13, 14]. For study of hybrids with another C57BL/6 strain harboring hemizygotic deletion of the nm23-m1 and nm23-m2 loci [m1m2]+/− [15], mice were crossed for at least 6 generations. Reduced NM23-M1 and NM23-M2 expression in the HGF+ × [m1m2]+/− hybrid relative to that of the wild-type C57BL/6 and HGF+ strains was verified in skin tissue by immunoblot analysis (Supplemental Fig. 1). Animals from backcrosses were further propagated to maintain the colony. To induce melanoma, neonatal male mice at postnatal day four were subjected to a single erythematous dose of UVB/A radiation (4,000 J/m2) in single wells of a 6-well plastic tissue culture plate without the lid. UVR was administered with lamps emitting a spectral output in the 290–400 nm range (72 % UVB, 27 % UVA, < 0.01 % UVC) (UVP, Upland, CA). Irradiated mice displayed skin reddening and occasional superficial desquamation, but without morbidity or mortality. Tumor incidence and growth rate (volume) were monitored weekly, with caliper measurements [16] initiated at first appearance of a raised lesion. Mice were utilized for the study and maintained for 10 months, but were sacrificed earlier if tumors reached a critical size of 500 mm3, or if overt signs of tumor-related illness were apparent. Necropsies were performed on all mice, and regional lymph nodes and internal organs were inspected for metastases. All skin lesions were analyzed by standard histopathology and immunohistochemical methods.
Establishment of murine melanoma cell lines
Melanoma cell cultures were established from melanomas located on back skin arising in both HGF+ and HGF+ × [m1m2]+/− mice using methods previously described [17, 18] (Supplemental Tables 1, 2). Briefly, hair surrounding the melanoma tumor was removed by a chemical depilatory method, followed by sterile excision of the lesion. Tumor tissue was washed with sterile PBS containing Anti–Anti (Invitrogen, Carlsbad, CA), cut with sterile cross blades to yield 1 mm sized pieces, and mixed with collagenase IV solution for 4 h at 4 °C to obtain single cell suspensions. The slurry was then incubated with trypsin–EDTA incubation overnight at 4 °C (Invitrogen, Carlsbad, CA). The resultant cell suspension was collected by centrifugation at 2,000×g for 5 min. Cell pellets were resuspended and plated on poly-d-lysine coated cell culture dishes in MCDB media (Sigma, St. Louis, MO) supplemented with 2 mM CaCl2, 2.5 μg insulin, cholera toxin (20 nM), TPA (20 nM), stem cell factor (20 nM), 10 % horse serum and 10 % fetal calf serum (Invitrogen) and incubated at 37 °C. Cell lines were maintained in this growth medium for two passages, after which they were maintained in MCDB media. The cell lines established from HGF+ were designated A-T1, A-T2 and A-T3, and those from the HGF+ × [m1m2]+/− named B-T2, B-T5 and B-T6 (Supplemental Tables 1, 2).
Wound healing (scratch) assays
Cell lines were plated onto 24-well plates (3 × 105 cells/well) and incubated in 5 % CO2 at 37 °C and grown to 100 % confluence. The cells were rendered quiescent by changing the medium to MCDB without FBS for 24 h. The wound was achieved by scratching across the center of each well with a 200 μl pipette tip followed by a PBS wash to remove debris. Videomicroscopy (Nixon Eclipse Ti, Melville, NY) was utilized to monitor cell lines over a 24 h period with images acquired every 30 min. Cell motility was determined by percent wound closure (calculated by dividing distance moved of invading front at the indicated time points) and mean speed of cell migration (calculated by dividing the total distance of migration by indicated time points, at least 20 cells per cell line were analyzed).
DNA damage and repair assays
Cell lines derived from HGF+ and HGF+ × [m1m2]+/− tumors were irradiated with either 10 J/m2 UVB/A following 24 h in reduced serum medium (0.5 %). Removal of pyrimidine (6–4) pyrimidone photoproducts (6–4 photoproducts) was measured by immunoslot blot assay [19]. DNA (100 ng) was isolated (Qiagen, Valencia, CA) heat-denatured at 100 °C for 10 min, applied to a Hybond nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ) using a vacuum-driven slot blot apparatus and fixed by baking for 1 h at 80 °C. Membranes were incubated with mouse monoclonal antibodies specific for 6–4 photoproducts (Cosmo Bio, Tokyo, Japan). Peroxidase-conjugated anti-mouse secondary antibody was used at a dilution of 1/10,000 in blocking buffer. Equal loading of DNA was confirmed by DAPI (Invitrogen, Carlsbad, CA) staining [20].
Mutation frequency in the hprt gene
Acquired resistance of cells to 6-thioguanine (6-TG) is conferred primarily by mutations within the hprt locus [21], and quantified as the number of 6-TG-resistant (6-TGr) colonies obtained after selection. Frequencies of spontaneous and UVR-induced hprt mutations in HGF+ and HGF+ × [m1m2]+/−-derived cell lines as described [21]. Cell lines were seeded at 100 cells per well in a 6-well plate, with each line plated with at least six replicates. Cells were exposed to either UVB/A (5 J/m2) or sham-treated, then grown in complete MCDB medium supplemented with 20 μM 6-TG [12]. Colonies were counted at 21 days following initial treatment, with colony-forming efficiency derived as the number of 6-TGr colonies as a percentage of initial plating density.
Results
A tandem hemizygous deletion of the nm23-m1 and nm23-m2 genes does not affect incidence or growth rate of UVR-induced melanomas in the HGF+ mouse
To assess the potential relationship between NM23 protein expression and melanoma progression, mice harboring a tandem, hemizygotic deletion of the nm23-m1 and nm23-m2 loci ([m1m2]+/− strain) [15] were crossed with an HGF-overexpressing strain (HGF+). The HGF+ strain is prone to UVR-induced melanoma with rapid growth characteristics but low metastatic potential [14, 22]. Newborn litters (post-natal day 4) from the parental strains and the HGF+ × [m1m2]+/− hybrid were exposed to UVR and monitored for development and growth of skin tumors (Fig. 1a). Tandem, homozygous deletion of the nm23-m1 and nm23-m2 genes is lethal in late embryonic and early neonatal development, preventing analysis of this condition. We have shown previously that UVR treatment of neonatal [m1m2]+/− mice induces relatively small nonmetastatic lesions consistent with melanoma in situ on exposed surfaces of the tail, but at no other locations [12]. Confinement of tumors to this location is likely due to the unique external (and UVR-accessible) location of tail melanocytes in the C57BL/6 mouse, which are more deeply sequestered in other skin areas within the dermis and are thus more protected from UVR. In that study, no large melanomas were obtained in the [m1m2]+/− strain on other UVR-exposed skin areas such as the back, ears, paws, etc.
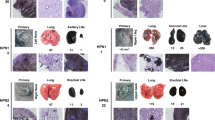
Consistent with prior reports [23, 24], UVR treatment elicited pigmented skin tumors in the majority (68 %) of HGF+ mice (Table 1), with no mice displaying more than one tumor. Most of the melanomas were located on the back skin (Fig. 1b, c), although some were also observed on other UVR-exposed locations such as the tail, ear and leg. While time of tumor onset was not significantly different between back and non-back tumors (~150 day; Fig. 2a, b), back tumors grew much more aggressively after their initial appearance (Fig. 2c,d; Supplemental Tables 1, 2).
A tandem hemizygous deletion of the nm23-m1 and nm23-m2 genes does not affect UVR-induced incidence or growth rate of melanomas in the HGF+ mouse. a Kaplan–Meier plot of back and non-back melanoma free survival and b mean age of back and non-back melanoma onset (days ± SEM). Tumor growth kinetics of c back melanomas (HGF+ (n = 12); HGF+ × [m1m2]+/− (n = 10) and d non-back melanomas [HGF+ (n = 5)]; HGF+ × [m1m2]+/− (n = 6)
Crossing of the [m1m2]+/− genotype into the HGF+ background had no effect on melanoma location (Fig. 1b; Table 1), incidence (Fig. 2a, b; Table 1), onset (Fig. 2a, b), grade or volume (Fig. 2c, d; Supplemental Tables 1, 2). The lack of effect of NM23 deficiency on growth of HGF+-driven melanomas is consistent with prototypical metastasis suppressor activity, in which metastatic potential but not primary tumor growth per se is impacted. The incidence of lower-grade, UVR-induced pigmented follicular cyst/pigmented epithelioid melanocytoma (PEM) tumors on tail skin was much lower in the HGF+ and HGF+ × [m1m2]+/− hybrid strains than the 100 % penetrance previously reported for the [m1m2]+/− strain [12]. This may be attributable to a more intense pigmentation conferred by the HGF+ genotype to tail skin of both the HGF+ and HGF+ [m1m2]+/− hybrid strains. In the absence of UVR, no skin tumors were observed in either the HGF+ or the HGF+ [m1m2]+/− hybrid groups, indicating the observed tumors were initiated by UVR treatment.
nm23-m1 and nm23-m2-deficiency induces metastasis of UVR-induced melanomas generated on back skin
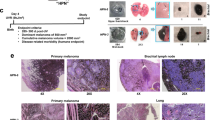
To measure the effect of NM23 deficiency on metastasis of UVR-induced melanoma, draining lymph nodes and visceral organs examined both grossly and microscopically for melanocyte infiltration. No macroscopic evidence of metastasis was associated with melanomas arising at any skin location for the HGF+ strain, consistent with the low metastatic potential described previously for this model system [22, 25]. In marked contrast, however, all melanomas arising on the back skin of HGF+ × [m1m2]+/− mice, independent of tumor size and growth rate, showed evidence of metastatic spread (Table 1). No evidence of metastasis was observed with the six slow-growing HGF+ × [m1m2]+/− melanomas/PEMs that all appeared on skin locations other than the back, which was likely related to their much slower growth and reduced primary tumor mass. Lymph node metastasis was seen in almost all back skin melanomas (9/10), while most also exhibited aggressive lung metastasis (7/10) with numerous pigmented nodules. Most of the affected lungs were also associated with multiple pigmented masses in the thoracic cavity (5/10), with metastases also seen in liver (4/10), and bone (1/10) (Fig. 3a, b; Supplemental Table 2). All of these sites are characteristic of melanoma metastasis in humans. In lymph nodes, metastatic growth was closely associated with node sinuses, strongly suggesting infiltration by melanoma cells occurred via lymphatic vessels (Fig. 3b). Moreover, lung and liver metastases were often seen in proximity to blood vessels, consistent with their entry into those organs via the circulatory system (Fig. 3b). In melanoma-bearing HGF+ mice, no microscopic evidence of pigmented micrometastases was evident in potential target organs for metastasis (data not shown).
Deficiency in nm23-m1 and nm23-m2-expression enhances UVR-induced melanoma metastasis. a Representative macroscopic appearance of metastases in lymph node, lung, thoracic cavity and liver scale bar represent 2 mm. b Representative H&E stained sections of common metastatic locations, scale bars represent 500 and 100 μm, respectively. Arrows indicate metastases found in proximity to lung and liver blood vessels. Note no metastatic dissemination was observed in the HGF+ transgenic mouse
Deficiency in nm23-m1 and nm23-m2 expression does not affect histopathological characteristics of UVR-induced primary back melanomas in the HGF+ mouse
UVR-induced melanomas of the HGF+ and HGF+ × [m1m2]+/− strains exhibited histopathological characteristics identical to those previously described for the HGF+ strain [26, 27]. The rapidly-growing back skin melanomas consisted of heavily pigmented epitheloid cells, arranged asymmetrically with numerous spindle-shaped cells and mitotic figures (Fig. 4a). In both the HGF+ and HGF+ × [m1m2]+/− groups, heavily pigmented cells filled and expanded the dermis, in most cases extending vertically through the subcutis, reminiscent of that found in CMM human patients [14, 27]. Extensive histological injury to the epidermal and dermal architecture (i.e. ulceration and destruction of hair follicles) was commonly evident. In both mouse groups, a slower-growing melanoma sub-type also was evident and presented with dermal aggregates of heavily pigmented epitheloid cells and subcutical invasion. However, these tumors were characterized by asymmetric vertical strands of tumor tissue, smaller contiguous tumor masses, and minimal intradermal expansion (Fig. 4b). Expression of NM23-M1 and NM23-M2 in primary melanomas of the HGF+ × [m1m2]+/− group remained at the expected 50 % reduced levels relative to those obtained in HGF+ mice (Supplemental Fig. 2). This verified that potential changes, such as compensatory increases or decreases (e.g. due to loss of heterozygosity) did not occur in HGF+ × [m1m2]+/− tumors and were not a possible underlying cause of their increased metastatic potential. The lack of a clear difference in overall invasive index between back skin melanomas of the two mouse groups, indicates that the high metastatic potential associated with the HGF+ × [m1m2]+/− genotype is not determined by overall tumor expansion outside the dermal margins.
Deficiency in nm23-m1 and nm23-m2 expression does not affect histopathological characteristics of UVR-induced primary back melanomas in the HGF+ mouse. Fast and slower-growing melanoma subtypes were observed. Representative H&E-stained sections of a rapidly growing back melanoma and b slower-growing back melanomas in HGF+ and HGF+ × [m1m2]+/− transgenic animals. Scale bars are indicated
Cell lines derived from back skin melanomas of the HGF+ × [m1m2]+/− strain exhibit increased cellular motility, impaired DNA repair capacity, and genomic instability
To measure the effect of NM23 expression on motility and genomic stability in melanomas of the HGF+ and HGF+ × [m1m2]+/− mouse strains, cell lines were established for more detailed analysis of these phenotypes. Three cell lines were successfully developed from separate back skin melanomas of the HGF+ strain (designated A-T1, A-T2 and A-T3) and HGF+ × [m1m2]+/− hybrid (B-T2, B-T5 and B-T6) (Supplemental Tables 1, 2). As expected, expression of NM23-M1 and NM23-M2 protein was reduced by approximately 50 % in cell lines established from HGF+ × [m1m2]+/− melanomas (Supplemental Fig. 3). Spontaneous cell motility was evaluated using the standard scratch/wounding assay in conditions of low mitogenicity culture medium. All three HGF+-derived cell lines consisted of cells that displayed minimal migration with short trajectories, in contrast to cells isolated from HGF+ × [m1m2]+/− melanomas that exhibited highly motile behavior and much longer trajectories (Fig. 5a). Furthermore, the HGF+-derived cell lines displayed a slower migration into a “scratch wound” over a 24 h time course, characterized mainly by slow movement of broad adherent cell masses and occasionally single cells (Fig. 5b; Supplemental Video Files 1–6). In addition, to the highly motile nature of HGF+ × [m1m2]+/− cells, they migrated primarily as detached single cells that travelled at a greater speed which almost completely filled a “scratch” wound within 12–24 h (Fig. 5b, c, d). These results suggest that enhanced single cell migration in NM23-deficient melanoma cell lines may underlie their high metastatic potential.
Melanoma cell lines derived from the HGF+ × [m1m2]+/− transgenic mouse exhibit increased cellular migratory response. Isolated melanoma-derived cell lines were tracked by videomicroscopy over a 24 h time period. a Migratory patterns of single cells established from HGF+ and HGF+ × [m1m2]+/− tumors during a 24 h period. b Representative photomicrographs of confluent monolayers at 0, 6, 12 and 24 h after wounding in HGF+ and HGF+ × [m1m2]+/− cell lines, scale bar represents 20 μm c percent closure of wound and d mean speed of migration. Values significantly different as determined by 2-way ANOVA (*P ≤ 0.05; n = 3; bars SEM)
To determine whether genomic stability was compromised in back skin melanomas of the HGF+ × [m1m2]+/− strain, DNA repair capacity and mutation rates were measured in the cell lines derived from each genotype. DNA repair capacity was measured in terms of the elimination of DNA damage in the form of UVB/A-induced 6–4 photoproducts using immuno-slot blot analysis, as previously described [12]. Nucleotide excision repair (NER) is the principal mechanism for repair of UVR-generated (6–4) photoproducts [28], and we recently demonstrated that NER capacity is reduced in nm23-deficient murine melanocytes and mouse embryonic fibroblasts [12]. In all three of the HGF+ × [m1m2]+/− cell lines (6–4) photoproduct repair was slower than that of HGF+ cell lines (t1/2 6.3 h vs t1/2 2.5 h; P ≤ 0.05) (Fig. 6a, b).
Melanoma cell lines derived from the HGF+ × [m1m2]+/− transgenic mouse exhibit compromised UVR-induced DNA repair and increased mutagenesis. DNA repair was measured by immuno-slot blot analysis following UVB/A exposure (10 J/m2). a Relative DNA repair efficiencies and b representative immuno-slot blot. c Spontaneous and d UVR-induced colony-forming efficiency was determined at 21 days post-6-TG treatment as described in “Materials and Methods”. Values significantly different as determined by 2-way ANOVA (*P ≤ 0.05; n = 3; bars SEM)
Rates of spontaneous and UVR-induced mutagenesis were quantified in the panel of melanoma cell lines using the 6-TGr colony formation assay [11]. In the absence of UVR exposure, spontaneous mutation rates (i.e. colony formation) were significantly higher in all three HGF+ × [m1m2]+/− cell lines compared to the HGF+ lines (2- to 3-fold; P ≤ 0.05) (Fig. 6c). UVB/A exposure caused an overall 2- to 3-fold increase in mutation rates across all six cell lines relative to unexposed cells, with the HGF+ × [m1m2]+/− cell lines retaining 2-fold higher rates compared to their HGF+ counterparts (P ≤ 0.05) (Fig. 6d). Taken together, the increases in single cell motility and genomic instability observed in cell lines from NM23-deficient melanomas identifies potential roles for both characteristics in driving the high metastatic potential of these tumors.
Discussion
The NM23-deficient mouse strain has provided important in vivo validation of the metastasis suppressor function of NM23 in melanoma. Hemizygosity in the nm23-m1 and nm23-m2 genes conferred strong metastatic activity to primary melanomas induced by UVR on back skin in the HGF+ mouse strain without effect on primary tumor growth characteristics. This is consistent with prototypical metastasis suppressor activity, and corroborates an earlier report of similar pro-metastatic effects of nm23-m1 deficiency in the context of virus-induced hepatocarcinoma [29]. Our study demonstrates an exquisite sensitivity of the HGF+-driven melanoma model, to modest 50 % reductions in NM23-M1 and NM23-M2 expression, with all back skin tumors exhibiting lymph node and/or organ metastasis. Since both the nm23-m1 and nm23-m2 genes were deleted concurrently, it remains to be determined whether one isoform exerts a dominant role or cooperativity exists between them. The Lacombe laboratory [30] has recently reported that silencing nm23-h1, but not nm23-h2, promotes the invasive phenotype in liver and colon carcinoma cell lines, strongly suggesting the H1/M1 isoform plays the dominant role in metastasis suppression. Transgenic mice with the m1 and m2 genes individually deleted are currently being crossed with the HGF+ strain to address this issue directly.
The lack of distinguishing histopathological characteristics associated with primary melanomas of the HGF+ × [m1m2]+/− strain was surprising in light of the strong pro-metastatic effect of NM23 deficiency. Although poorly metastatic, HGF+ melanomas grow aggressively and often invade en masse into the subcutical layer of skin, suggesting metastasis is blocked at a later stage of the metastatic cascade (e.g. intravasation, survival in the circulation, extravasation or colonization/angiogenesis). The enhanced ability of HGF+ × [m1m2]+/−- derived melanoma cell lines to migrate autonomously (i.e. in the absence of exogenous chemoattractant) as single cells in scratch assays suggests a clue to the metastatic behavior of these tumors. Such activity could promote migration and invasion toward blood and lymphatic vessels, as well as their traversing of vessel walls at primary and metastatic sites.
While the melanoma-derived cell lines derived from HGF+ × [m1m2]+/− mice exhibited greater motility/invasion and genomic instability than their HGF+-derived counterparts, it remains to be determined whether one or both of these phenotypes are metastasis-suppressing functions. Complementation of the mouse genotype with NM23 mutants that selectively disrupt the motility-suppressing and genome-stabilizing functions of the molecule could offer insights to this key question. Metastasis-prone melanomas of the HGF+ × [m1m2]+/− strain are likely to have acquired metastasis-driving mutations, and loss of the genomic stabilizing activity of NM23 is a plausible candidate mechanism. NM23 deficiency only impacted the metastatic phenotype and not primary tumor growth (i.e. incidence and melanoma onset), suggesting effects of NM23-dependent genomic instability may be selectively directed to metastasis-driving and not tumor-driving mutations. However, the robust nature of UVR-induced melanomas obtained in the HGF+ model strongly suggests tumor-driving mutations are readily acquired independent of NM23 status, which may have obscured the effects of nm23-m1/m2 deficiency on tumor initiation and growth [12]. Taken together, our findings suggest the dual anti-motility and genomic stabilizing activities of NM23 could provide a synergistic boost needed to overcome barriers to metastatic growth.
BRAF mutations at the V600 residue are the predominant MAPK activating mutations found in cutaneous melanoma. Such mutations are rarely if ever seen in the HGF mouse melanoma model [24, 31, 32], which is probably a consequence of constitutive RAF activation downstream of the cell surface receptor for HGF, the MET tyrosine kinase. The ability of NM23 deficiency to confer metastatic potential in HGF-driven melanoma suggests the HGF+ × [m1m2]+/− strain may be a relevant experimental model for human melanomas that do not harbor BRAF mutations (20–40 %) and are unsuitable candidates for anti-BRAF therapies [33]. Efforts are underway to identify metastasis-driving mutations and other molecular events in NM23-deficient melanomas of these transgenic knockout mouse strains.
Abbreviations
- CMM:
-
Cutaneous malignant melanoma
- UVR:
-
Ultraviolet radiation
- NDPK:
-
Nucleoside diphosphate kinase
- HisK:
-
Histidine kinase
- HGF:
-
Hepatocyte growth factor
- MAPK:
-
Mitogen-activated protein kinase
- NER:
-
Nucleotide excision repair
- PEM:
-
Pigmented epithelioid melanocytoma
References
Linos K, Slominski A, Ross JS, Carlson JA (2011) Melanoma update: diagnostic and prognostic factors that can effectively shape and personalize management. Biomark Med 5(3):333–360
Gandini S, Autier P, Boniol M (2011) Reviews on sun exposure and artificial light and melanoma. Prog Biophys Mol Biol 107(3):362–366
Green AC, Wallingford SC, McBride P (2011) Childhood exposure to ultraviolet radiation and harmful skin effects: epidemiological evidence. Prog Biophys Mol Biol 107(3):349–355
Steeg PS, Bevilacqua G, Pozzatti R, Liotta LA, Sobel ME (1988) Altered expression of NM23, a gene associated with low tumor metastatic potential, during adenovirus 2 Ela inhibition of experimental metastasis. Cancer Res 48(22):6550–6554
Leone A, Flatow U, King CR, Sandeen MA, Margulies IM, Liotta LA, Steeg PS (1991) Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 65(1):25–35
Florenes VA, Aamdal S, Myklebost O, Maelandsmo GM, Bruland OS, Fodstad O (1992) Levels of nm23 messenger RNA in metastatic malignant melanomas: inverse correlation to disease progression. Cancer Res 52(21):6088–6091
Ferrari D, Lombardi M, Ricci R, Michiara M, Santini M, De Panfilis G (2007) Dermatopathological indicators of poor melanoma prognosis are significantly inversely correlated with the expression of NM23 protein in primary cutaneous melanoma. J Cutan Pathol 34(9):705–712
Agarwal RP, Robison B, Parks RE Jr (1978) Nucleoside diphosphokinase from human erythrocytes. Methods Enzymol 51:376–386
Freije JM, Blay P, MacDonald NJ, Manrow RE, Steeg PS (1997) Site-directed mutation of Nm23-H1. Mutations lacking motility suppressive capacity upon transfection are deficient in histidine-dependent protein phosphotransferase pathways in vitro. J Biol Chem 272(9):5525–5532
Ma D, McCorkle JR, Kaetzel DM (2004) The metastasis suppressor NM23-H1 possesses 3′–5′ exonuclease activity. J Biol Chem 279(17):18073–18084
Yang M, Jarrett SG, Craven R, Kaetzel DM (2009) YNK1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation- and etoposide-induced DNA damage. Mutat Res 660(1–2):74–78
Jarrett SG, Novak M, Dabernat S, Daniel JY, Mellon I, Zhang Q, Harris N, Ciesielski MJ, Fenstermaker RA, Kovacic D et al (2012) Metastasis suppressor NM23-H1 promotes repair of UV-induced DNA damage and suppresses UV-induced melanomagenesis. Cancer Res 72(1):133–143
Otsuka T, Takayama H, Sharp R, Celli G, LaRochelle WJ, Bottaro DP, Ellmore N, Vieira W, Owens JW, Anver M et al (1998) c-Met autocrine activation induces development of malignant melanoma and acquisition of the metastatic phenotype. Cancer Res 58(22):5157–5167
Noonan FP, Otsuka T, Bang S, Anver MR, Merlino G (2000) Accelerated ultraviolet radiation-induced carcinogenesis in hepatocyte growth factor/scatter factor transgenic mice. Cancer Res 60(14):3738–3743
Postel EH, Wohlman I, Zou X, Juan T, Sun N, D′Agostin D, Cuellar M, Choi T, Notterman DA, La Perle KM (2009) Targeted deletion of Nm23/nucleoside diphosphate kinase A and B reveals their requirement for definitive erythropoiesis in the mouse embryo. Dev Dyn 238(3):775–787
Tomayko MM, Reynolds CP (1989) Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol 24(3):148–154
Scott TL, Wakamatsu K, Ito S, D’Orazio JA (2009) Purification and growth of melanocortin 1 receptor (Mc1r)-defective primary murine melanocytes is dependent on stem cell factor (SFC) from keratinocyte-conditioned media. In Vitro Cell Dev Biol Anim 45(10):577–583
Soo JK, Ross AD, Bennett DC (2011) Isolation and culture of melanoma and naevus cells and cell lines. Methods Mol Biol 731:141–150
Mellon I, Hock T, Reid R, Porter PC, States JC (2002) Polymorphisms in the human xeroderma pigmentosum group A gene and their impact on cell survival and nucleotide excision repair. DNA Repair 1(7):531–546
Wulff BC, Schick JS, Thomas-Ahner JM, Kusewitt DF, Yarosh DB, Oberyszyn TM (2008) Topical treatment with OGG1 enzyme affects UVB-induced skin carcinogenesis. Photochem Photobiol 84(2):317–321
Glaab WE, Tindall KR (1997) Mutation rate at the hprt locus in human cancer cell lines with specific mismatch repair-gene defects. Carcinogenesis 18(1):1–8
Takayama H, La Rochelle WJ, Sharp R, Otsuka T, Kriebel P, Anver M, Aaronson SA, Merlino G (1997) Diverse tumorigenesis associated with aberrant development in mice overexpressing hepatocyte growth factor/scatter factor. Proc Natl Acad Sci USA 94(2):701–706
Landsberg J, Gaffal E, Cron M, Kohlmeyer J, Renn M, Tuting T (2010) Autochthonous primary and metastatic melanomas in Hgf-Cdk4 R24C mice evade T-cell-mediated immune surveillance. Pigment Cell Melanoma Res 23(5):649–660
Gaffal E, Landsberg J, Bald T, Sporleder A, Kohlmeyer J, Tuting T (2011) Neonatal UVB exposure accelerates melanoma growth and enhances distant metastases in Hgf-Cdk4(R24C) C57BL/6 mice. Int J Cancer 129(2):285–294
De Fabo EC, Noonan FP, Fears T, Merlino G (2004) Ultraviolet B but not ultraviolet A radiation initiates melanoma. Cancer Res 64(18):6372–6376
Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL, De Fabo EC, Merlino G (2001) Neonatal sunburn and melanoma in mice. Nature 413(6853):271–272
Florell SR, Thomas J, Grossman D (2007) Predominant formation of heavily pigmented dermal melanocytomas resembling ‘animal-type’ melanomas in hepatocyte growth factor (C57BL/6 × C3H)F1 mice following neonatal UV irradiation. J Cutan Pathol 34(9):667–674
Shuck SC, Short EA, Turchi JJ (2008) Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res 18(1):64–72
Boissan M, Wendum D, Arnaud-Dabernat S, Munier A, Debray M, Lascu I, Daniel JY, Lacombe ML (2005) Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst 97(11):836–845
Boissan M, De Wever O, Lizarraga F, Wendum D, Poincloux R, Chignard N, Desbois-Mouthon C, Dufour S, Nawrocki-Raby B, Birembaut P et al (2010) Implication of metastasis suppressor NM23-H1 in maintaining adherens junctions and limiting the invasive potential of human cancer cells. Cancer Res 70(19):7710–7722
Recio JA, Noonan FP, Takayama H, Anver MR, Duray P, Rush WL, Lindner G, De Fabo EC, DePinho RA, Merlino G (2002) Ink4a/arf deficiency promotes ultraviolet radiation-induced melanomagenesis. Cancer Res 62(22):6724–6730
Noonan FP, Dudek J, Merlino G, De Fabo EC (2003) Animal models of melanoma: an HGF/SF transgenic mouse model may facilitate experimental access to UV initiating events. Pigment Cell Res 16(1):16–25
Garnett MJ, Marais R (2004) Guilty as charged: B-RAF is a human oncogene. Cancer Cell 6(4):313–319
Acknowledgments
The authors wish to thank Edith Postel for providing the [m1m2]+/− mouse strain, and John D’Orazio and Kathryn Coyle for their helpful advice over the course of these studies. This work was supported by the National Institutes of Health, National Cancer Institute Grant CA83237 (D.M.K).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 8 (AVI 203687 kb)
Supplementary material 9 (AVI 203687 kb)
Supplementary material 10 (AVI 203687 kb)
Supplementary material 11 (AVI 203687 kb)
Supplementary material 12 (AVI 203687 kb)
Supplementary material 13 (AVI 203687 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jarrett, S.G., Novak, M., Harris, N. et al. NM23 deficiency promotes metastasis in a UV radiation-induced mouse model of human melanoma. Clin Exp Metastasis 30, 25–36 (2013). https://doi.org/10.1007/s10585-012-9495-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-012-9495-z