Abstract
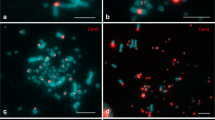
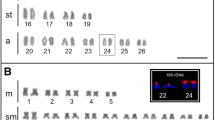
Many families of centromeric repetitive DNA sequences isolated from Struthioniformes, Galliformes, Falconiformes, and Passeriformes are localized primarily to microchromosomes. However, it is unclear whether chromosome size-correlated homogenization is a common characteristic of centromeric repetitive sequences in Aves. New World and Old World quails have the typical avian karyotype comprising chromosomes of two distinct sizes, and C-positive heterochromatin is distributed in centromeric regions of most autosomes and the whole W chromosome. We isolated six types of centromeric repetitive sequences from three New World quail species (Colinus virginianus, CVI; Callipepla californica, CCA; and Callipepla squamata, CSQ; Odontophoridae) and one Old World quail species (Alectoris chukar, ACH; Phasianidae), and characterized the sequences by nucleotide sequencing, chromosome in situ hybridization, and filter hybridization. The 385-bp CVI-MspI, 591-bp CCA-BamHI, 582-bp CSQ-BamHI, and 366-bp ACH-Sau3AI fragments exhibited tandem arrays of the monomer unit, and the 224-bp CVI-HaeIII and 135-bp CCA-HaeIII fragments were composed of minisatellite-like and microsatellite-like repeats, respectively. ACH-Sau3AI was a homolog of the chicken nuclear membrane repeat sequence, whose homologs are common in Phasianidae. CVI-MspI, CCA-BamHI, and CSQ-BamHI showed high homology and were specific to the Odontophoridae. CVI-MspI was localized to microchromosomes, whereas CVI-HaeIII, CCA-BamHI, and CSQ-BamHI were mapped to almost all chromosomes. CCA-HaeIII was localized to five pairs of macrochromosomes and most microchromosomes. ACH-Sau3AI was distributed in three pairs of macrochromosomes and all microchromosomes. Centromeric repetitive sequences may be homogenized in chromosome size-correlated and -uncorrelated manners in New World quails, although there may be a mechanism that causes homogenization of centromeric repetitive sequences primarily between microchromosomes, which is commonly observed in phasianid birds.










Similar content being viewed by others
Abbreviations
- AAU:
-
Amazona autumnalis
- ACH:
-
Alectoris chukar
- ACY:
-
Anser cygnoides var. orientalis
- APL:
-
Anas platyrhynchos var. domesticus
- BBL:
-
Bubo blakistoni
- BrdU:
-
5-Bromo-2′-deoxyuridine
- CCA:
-
Callipepla californica
- CCD:
-
Charge-coupled device
- CCH:
-
Coturnix chinensis
- CCY:
-
Cygnus cygnus
- cDNA:
-
Complementary DNA
- CJA:
-
Coturnix japonica
- CNM:
-
Chicken nuclear membrane
- CSQ:
-
Callipepla squamata
- CVI :
-
Colinus virginianus
- DDBJ:
-
DNA Data Bank of Japan
- DIG:
-
Digoxigenin
- DNO:
-
Dromaius novaehollandiae
- dNTP:
-
Deoxynucleotide triphosphate
- EEL:
-
Eudromia elegans
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- GGA:
-
Gallus gallus
- GLE:
-
Grus leucogeranus
- HRU:
-
Hirundo rustica
- Kb:
-
Kilobase pairs
- NME:
-
Numida meleagris
- NNI:
-
Nisaetus nipalensis orientalis
- PAT:
-
Probosciger aterrimus
- PCR:
-
Polymerase chain reaction
- PHA:
-
Pandion haliaetus
- PI:
-
Propidium iodide
- PMI:
-
Platalea minor
- SCA:
-
Struthio camelus
- SDS:
-
Sodium dodecyl sulfate
- SSC:
-
Saline sodium citrate
- TM:
-
Turkey microchromosome
- UV:
-
Ultraviolet
References
Burrack LS, Berman J (2012) Flexibility of centromere and kinetochore structures. Trends Genet 28:204–212
Chen ZQ, Lin CC, Hodgetts RB (1989) Cloning and characterization of a tandemly repeated DNA sequence in the crane family (Gruidae). Genome 32:646–654
Christidis L (1990) Animal cytogenetics 4: Chordata 3 B: Aves. Gebrüder Borntraeger, Berlin
de Boer LEM (1976) The somatic chromosome complements of 16 species of Falconiformes (Aves) and the karyological relationships of the order. Genetica 46:77–113
de Boer LEM, Sinoo RP (1984) A karyological study of Accipitridae (Aves: Falconiformes), with karyotypic descriptions of 16 species new to cytology. Genetica 65:89–107
de la Sena CA, Nestor KE (1991) Variability of C-banding patterns in Japanese quail chromosomes. Genome 34:993–997
Griffin DK, Haberman F, Masabanda J et al (1999) Micro- and macrochromosome paints generated by flow cytometry and microdissection: tools for mapping the chicken genome. Cytogenet Cell Genet 87:278–281
Habermann FA, Cremer M, Walter J et al (2001) Arrangements of macro- and microchromosomes in chicken cells. Chromosomes Res 9:569–584
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
International Chicken Genome Sequencing Consortium (ICGSC) (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Kalitis P, Choo KHA (1997) Centromere DNA of higher eukaryotes. In: Choo KHA (ed) The centromere. Oxford University Press, Oxford, pp. 97–142
Kodama H, Saitoh H, Tone M, Kuhara S, Sakaki Y, Mizuno S (1987) Nucleotide sequences and unusual electrophoretic behavior of the W chromosome-specific repeating DNA units of the domestic fowl, Gallus gallus domesticus. Chromosoma 96:18–25
Longmire JL, Lewis AK, Brown NC et al (1988) Isolation and molecular characterization of a highly polymorphic centromeric tandem repeat in the family Falconidae. Genomics 2:14–24
Matsuda Y, Chapman VM (1995) Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16:261–272
Matzke MA, Varga F, Berger H et al (1990) A 41–42 bp tandemly repeated sequence isolated from nuclear envelopes of chicken erythrocytes is located predominantly on microchromosomes. Chromosoma 99:131–137
Matzke AJ, Varga F, Gruendler P et al (1992) Characterization of a new repetitive sequence that is enriched on microchromosomes of turkey. Chromosoma 102:9–14
Nishida C, Ishijima J, Ishishita S, Yamada K, Griffin DK, Yamazaki T, Matsuda Y (2013a) Karyotype reorganization with conserved genomic compartmentalization in dot-shaped microchromosomes in the Japanese mountain hawk-eagle (Nisaetus nipalensis orientalis, Accipitridae). Cytogenet Genome Res 141:284–294
Nishida C, Ishishita S, Yamada K, Griffin DK, Matsuda Y (2013b) Dynamic chromosome reorganization in the osprey (Pandion haliaetus, Pandionidae, Falconiformes): relationship between chromosome size and the chromosomal distribution of centromeric repetitive DNA sequences. Cytogenet Genome Res (In press)
Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y, Griffin DK (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res 15:721–734
Saifitdinova AF, Derjusheva SE, Malykh AG, Zhurov VG, Andreeva TF, Gaginskaya ER (2001) Centromeric tandem repeat from the chaffinch genome: isolation and molecular characterization. Genome 44:96–103
Saitoh H, Harata M, Mizuno S (1989) Presence of female-specific bent-repetitive DNA sequences in the genomes of turkey and pheasant and their interactions with W-protein of chicken. Chromosoma 98:250–258
Sasaki M, Nishida C (1980) C-banded heteromorphism in the Z chromosome of the Japanese quail, Coturnix c. japonica. Chrom Inf Serv 29:21–22
Shang WH, Hori T, Toyoda A et al (2010) Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res 20:1219–1228
Shibusawa M, Nishida-Umehara C, Tsudzuki M, Masabanda J, Griffin DK, Matsuda Y (2004a) A comparative karyological study of the blue-breasted quail (Coturnix chinensis, Phasianidae) and California quail (Callipepla californica, Odontophoridae). Cytogenet Genome Res 106:82–90
Shibusawa M, Nishibori M, Nishida-Umehara C et al (2004b) Karyotypic evolution in the Galliformes: an examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenet Genome Res 106:111–119
Shoffner RN (1974) Chromosomes in birds. In: Busch H (ed) The cell nucleus. Academic, San Diego, pp. 223–261
Singer MF (1982) Highly repeated sequences in mammalian genomes. Int Rev Cytol 76:67–112
Solovei IV, Joffe BI, Gaginskaya ER, Macgregor HC (1996) Transcription of lampbrush chromosomes of a centromerically localized highly repeated DNA in pigeon (Columba) relates to sequence arrangement. Chromosome Res 4:588–603
Stock AD, Bunch TD (1982) The evolutionary implications of chromosome banding pattern homologies in the bird order Galliformes. Cytogenet Cell Genet 34:136–148
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306
Suzuki T, Kurosaki T, Shimada K et al (1999) Cytogenetic mapping of 31 functional genes on chicken chromosomes by direct R-banding FISH. Cytogenet Cell Genet 87:32–40
Takagi N, Sasaki M (1974) A phylogenetic study of bird karyotypes. Chromosoma 46:91–120
Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T (2002) Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat Res 504:37–45
Tanaka K, Suzuki T, Nojiri T, Yamagata T, Namikawa T, Matsuda Y (2000) Characterization and chromosomal distribution of a novel satellite DNA sequence of Japanese quail (Coturnix coturnix japonica). J Hered 91:412–415
Uno Y, Nishida C, Tarui H et al (2012) Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS ONE 7:e53027
Yamada K, Nishida-Umehara C, Matsuda Y (2002a) Characterization and chromosomal distribution of novel satellite DNA sequences of the lesser rhea (Pterocnemia pennata) and the greater rhea (Rhea americana). Chromosome Res 10:513–523
Yamada K, Shibusawa M, Tsudzuki M, Matsuda Y (2002b) Molecular cloning and characterization of novel centromeric repetitive DNA sequences in the blue-breasted quail (Coturnix chinensis, Galliformes). Cytogenet Genome Res 98:255–261
Yamada K, Nishida-Umehara C, Matsuda Y (2004) A new family of satellite DNA sequences as a major component of centromeric heterochromatin in owls (Strigiformes). Chromosoma 112:277–287
Acknowledgments
This research was partially supported by the National BioResource Project (NBRP) Chicken/Quail, and Grants-in-Aid for Scientific Research on Innovative Areas (no. 23113004) and a Grant-in-Aid for Scientific Research (B) (no. 22370081) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: Darren K. Griffin and Beth A. Sullivan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Summary of copy numbers and amounts of repetitive sequences in the genome (XLSX 11 kb)
Supplementary Fig. 1
Nucleotide sequences of CVI-MspI, CVI-NsiI, and CVI-BamHI fragments. Asterisks indicate that sequences are rearranged to compare with CVI-BamHI. Dots indicate identity with nucleotides in the consensus sequence at the top, and hyphens indicate gaps. Internal restriction sites of six endonucleases are represented as follows: EcoRI ( ), BamHI (GGATCC), HpaII/MspI (CCGG), NsiI (
), BamHI (GGATCC), HpaII/MspI (CCGG), NsiI ( ), and TaqI (
), and TaqI ( ) (JPEG 1882 kb)
) (JPEG 1882 kb)
Supplementary Fig. 2
Nucleotide sequences of CCA-BamHI fragments. Dots indicate identity with nucleotides in the consensus sequence at the top, and hyphens indicate gaps. Restriction sites of three endonucleases are represented as follows: BamHI ( ), HpaII/MspI (CCGG), and TaqI (
), HpaII/MspI (CCGG), and TaqI ( ) (JPEG 1900 kb)
) (JPEG 1900 kb)
Supplementary Fig. 3
Nucleotide sequences of CSQ-BamHI fragments. Dots in CSQ-BamHI-33 indicate identity with nucleotides of CSQ-BamHI-26, and hyphens indicate gaps. Restriction sites of four endonucleases are represented as follows: BamHI ( ), HpaII/MspI (CCGG), TaqI (
), HpaII/MspI (CCGG), TaqI ( ), and HaeIII (GGCC) (JPEG 1862 kb)
), and HaeIII (GGCC) (JPEG 1862 kb)
Supplementary Fig. 4
Comparison of the CCA-BamHI consensus sequence, CSQ-BamHI-26 fragment (CSQ-BamHI), and the consensus sequences of the CVI-MspI, CVI-NsiI, and CVI-BamHI fragments (CVI-MspI). a Conserved regions are located at positions 2–334 bp and 547–589 bp of CCA-BamHI, which are indicated by dotted arrows. b Neighbor-joining tree representing phylogenetic relationships among the CCA-BamHI, CSQ-BamHI, and CVI-MspI sequence families (JPEG 1896 kb)
Supplementary Fig. 5
Southern blot hybridization patterns of three families of centromeric repetitive sequences, CVI-HaeIII, CCA-HaeIII, and ACH-Sau3AI. a Hybridization probed with the CVI-HaeIII-1 fragment to genomic DNA of C. virginianus female, which was digested with six endonucleases (HaeIII, BamHI, EcoRI, PstI, HpaII, and MspI). b Hybridization of the CCA-HaeIII-1 fragment to genomic DNA of C. californica female. c Hybridization of the ACH-Sau3AI-34 fragment to genomic DNA of A. chukar female. A mixture of λDNA–HindIII and φX174 DNA–HaeIII digests was used as a molecular weight marker (JPEG 1839 kb)
Supplementary Fig. 6
Comparison of southern blot hybridization patterns of CVI-MspI between male and female genomic DNA. Hybridization probed with the CVI-MspI-2 fragment to genomic DNA of C. virginianus male and female, which were digested with six endonucleases (MspI, HpaII, BamHI, EcoRI, NsiI, and PstI). A mixture of λDNA–HindIII and φX174 DNA–HaeIII digests was used as a molecular weight marker (JPEG 1831 kb)
Supplementary Fig. 7
Quantification of the CVI-HaeIII, CVI-MspI (CVI-NsiI, CVI-BamHI), CCA-BamHI, CCA-HaeIII, CSQ-BamHI, and ACH-Sau3AI family repetitive sequences in the genome by slot-blot hybridization. Eight different concentrations of genomic DNA of C. virginianus, C. californica, C. squamata, and A. chukar and plasmid DNA clones containing the DNA fragment of CVI-HaeIII-1, CVI-MspI-2, CCA-BamHI-1, CCA-HaeIII-1, CSQ-BamHI-26, and ACH-Sau3AI-34 were blotted on a membrane and probed by hybridization with the DNA fragment of CVI-HaeIII-1, CVI-MspI-2, CCA-BamHI-1, CCA-HaeIII-1, CSQ-BamHI-26, and ACH-Sau3AI-34 labeled with digoxigenin-11-dUTP (JPEG 2523 kb)
Rights and permissions
About this article
Cite this article
Ishishita, S., Tsuruta, Y., Uno, Y. et al. Chromosome size-correlated and chromosome size-uncorrelated homogenization of centromeric repetitive sequences in New World quails. Chromosome Res 22, 15–34 (2014). https://doi.org/10.1007/s10577-014-9402-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-014-9402-3