Abstract
Recently, a novel DNA replication precursor analogue called 5-ethynyl-2′-deoxyuridine (EdU) has been widely used to monitor DNA synthesis as an alternative to bromodeoxyuridine. Use of EdU benefits from simplicity and reproducibility and the simple chemical detection systems allows excellent preservation of nuclear structure. However, the alkyne moiety is highly reactive, raising the possibility that incorporation might compromise genome stability. To assess the extent of possible DNA damage, we have analysed the effect of EdU incorporation into DNA during short- and long-term cell culture using a variety of cell lines. We show that EdU incorporation has no measurable impact on the rate of elongation of replication forks during synthesis. However, using different cell lines we find that during long-term cell culture variable responses to EdU incorporation are seen, which range from delayed cell cycle progression to complete cell cycle arrest. The most profound phenotypes were seen in mouse embryonic stem cells, which following incorporation of EdU accumulated in the G2/M-phase of the cell cycle before undergoing apoptosis. In long-term cell culture, EdU incorporation also triggered a DNA damage response in all cell types analysed. Our study shows that while EdU is extremely useful to tag sites of on-going replication, for long-term studies (i.e. beyond the cell cycle in which labelling is performed), a careful analysis of cell cycle perturbations must be performed in order to ensure that any conclusions made after EdU treatment are not a direct consequence of EdU-dependent activation of cell stress responses.
Similar content being viewed by others
Introduction
In proliferating mammalian cells, different cell cycle stages are highly regulated in order to preserve genome integrity. In understanding the coordination of the different cell cycle stages, many experiments have relied on the use of DNA replication precursor analogues to monitor the replication process in situ and map the progression of the replicated DNA into mitosis and following cell cycles (Walter et al. 2003; Maya-Mendoza et al. 2010; Olivares-Chauvet et al. 2011). Numerous modified precursors have been used to label both sites of DNA synthesis and the distribution of the labelled sites in subsequent cell generations. Historically, 3H-thymidine was the first precursor in common use. This modified precursor, when added to culture medium and assimilated into the corresponding deoxyribonucleoside triphosphate was used to define critical features of the replication process, such as replication fork rate and the distribution of sites of initiation throughout S-phase (Fakan et al. 1972; Edenberg 1976; Blow et al. 2001). Soon after, bromodeoxyuridine (BrdU) was developed as a precursor of DNA synthesis and facilitated the visualisation of sites of DNA synthesis in nuclei of mammalian cells (Nakamura et al. 1986) and purification of regions of the genome that were replicated at specific times of S-phase (Farkash-Amar and Simon 2010). Subsequently, numerous precursors analogues coupled to biotin, digoxigenin and a wide range of fluorescent molecules have become available to study DNA replication (Philimonenko et al. 2006); the structure of the replication programme and the labelled sites and corresponding chromosomes both at the time of labelling and in subsequent cell cycles (Maya-Mendoza et al. 2009).
In recent years, a novel precursor analogue called ethynyldeoxyuridine (EdU) has been widely used as an alternative to BrdU to monitor DNA synthesis (Salic and Mitchison 2008). The major benefit of using EdU is in its simplicity: the reagent is added to medium, taken up by cells and incorporated into DNA, just as BrdU. However, whereas antibody based detection of BrdU incorporation requires denaturation of DNA for detection to work EdU is revealed by simple ‘Click’ chemistry, which is both efficient and reproducible. Hence, EdU incorporation and the corresponding fluorescent signal show a strong linear relationship, whereas antibody based detection of BrdU is non-linear as clusters of adjacent BrdU moieties cannot all be visualised because of steric hindrance during antibody binding. In addition to the relationship between EdU incorporation and fluorescent signal, the ease of EdU detection and enhanced preservation of nuclear structure has made EdU a reagent of choice for labelling sites of DNA synthesis (Buck et al. 2008).
In many published studies, EdU has been used to monitor S-phase progression and trace labelled cells in subsequent cell cycles (Salic and Mitchison 2008; Petruk et al. 2012; Aparicio et al. 2012). However, in recent years it has been reported that in EdU-labelled cells a variety of cell-type-specific changes in cell cycle behaviour occurs that demand careful analysis before EdU is adopted for routine use in long-term cell culture (Meneni et al. 2007; Diermeier-Daucher et al. 2009; Ross et al. 2011). To address this concern, we used a range of cell types that have not been studied before in this context, to analyse the impact of EdU on replication at the time of incorporation and then monitored the passage of the labelled cells into the next and subsequent cell cycles. We demonstrate that while EdU has little or no impact on the rate of elongation of replication forks during synthesis, mouse embryonic stem cells (mESC), in particular, show a subsequent accumulation and arrest in the G2/M-phase of the cell cycle. More notably, we find that mESC have very limited capacity for progression after EdU incorporation and die after the arrest. The response of different cell lines to EdU incorporation was shown to reflect the extent of activation of the DNA damage response (DDR) in these different cell types. Our study shows that while EdU is extremely useful to tag sites of on-going replication, for long-term studies (i.e. beyond the cell cycle in which labelling is performed) a careful analysis of cell cycle perturbations must be performed; this is necessary to ensure that any conclusions made after EdU treatment are not a direct consequence of EdU-dependent activation of cell stress responses.
Materials and methods
Cell culture
MESC (JM8A3.N1) were cultured in knockout Dulbecco’s modified Eagle’s medium (Gibco Invitrogen, Paisley, Renfrewshire, UK), supplemented with 10 % foetal bovine serum (FBS; Sigma-Aldrich Company, Dorset, UK), 1× l-glutamine (PAA Laboratories, Yeovil, Somerset, UK), 0.1 mM 2-mercaptoethanol (Sigma-Aldrich Company), 1× penicillin/streptomycin glutamine (Gibco Invitrogen) and 1,000 U/ml mouse leukaemia Inhibitory factor (Merck Millipore, Watford, Hertfordshire, UK), on 0.1 % (v/v) gelatine-coated (Sigma-Aldrich Company) 100-mm dishes (Corning, Birmingham, West Midlands, UK). Medium was changed every day, and cells were passaged onto a new dish every 3 days using 500,000–1,000,000 cells/dish.
Diploid human fibroblasts (hFb; MRC5) were grown in minimum essential medium (Sigma-Aldrich Company), supplemented with 10 % FBS, 1× non-essential amino acids (Sigma-Aldrich Company), 1× l-glutamine (Gibco Invitrogen) and 1× penicillin/streptomycin (Gibco Invitrogen) in 75 cm2 cell culture flasks (Corning). Cells were passaged into a new flask every 2–3 days using 1:3–1:6 dilution.
Mouse embryonic fibroblasts (MEFs) were cultured in DMEM (Lonza, Wolverhampton, UK), supplemented with 10 % FBS, 1× l-glutamine, 1× penicillin/streptomycin and 5 μM 2-mercaptoethanol on 100-mm dishes, and HeLa cells were grown in DMEM (Sigma-Aldrich Company), supplemented with 10 % FBS, 1× non-essential amino acids, 1× l-glutamine and 1× penicillin/streptomycin. MEF and HeLa cells were passaged onto a new dish every 2–3 days using 1:10–1:20 dilution. All cells types were grown at 37 °C in a 5 % (v/v) CO2 incubator.
Microscopy
Exponentially growing cells plated on 13-mm glass coverslips (VWR International, Lutterworthor, UK) were labelled with 10 μM BrdU (Sigma-Aldrich Company) or 10 μM EdU (Molecular Probes Invitrogen, Paisley, Renfrewshire, UK).
For pulse–chase–pulse labelling, cells were labelled with BrdU or EdU for 30 min, rinsed twice with warm medium, chased in fresh medium for 4 h, rinsed with ice-cold phosphate-buffered saline (PBS) and then transfected with 32 μM Cy3-dUTP (GE Healthcare UK, Little Chalfont, Buckinghamshire, UK) for 9 min on ice using Fugene6 (Roche Diagnostics, Burgess Hill, West Sussex, UK). After transfection, cells were rinsed with ice-cold PBS, then grown in fresh medium for 30 min, and fixed and stained as described below. For chromosome territory labelling, cells were labelled with BrdU or EdU for 30 min or 24 h, rinsed twice with warm medium, chased in fresh medium for 3 (mESC) or 5 (hFB) days, and then fixed and stained as described below. For DDR staining, cells were labelled with BrdU or EdU for 30 min (mESC) or 24 h (hFb), rinsed twice with warm medium, chased in fresh medium for 24 h, and then fixed and stained. After labelling, cells were fixed in 4 % paraformaldehyde (Electron Microscopy Sciences, Hatfield, UK) for 10 min and permeabilised with 1 % Triton X-100 (Sigma-Aldrich Company) for 10 min or 30 min for BrdU staining.
For EdU staining the Click-iT Alexa Fluor 488 Imaging Kit (Molecular Probes Invitrogen) was used following the manufacturer’s instructions. Cells were rinsed twice with 3 % bovine serum albumin (BSA; Sigma-Aldrich Company) and incubated with 150 μl EdU-Click reaction-mix (15 μl 10× reaction buffer, 6 μl CuSO4, 1.5 μl Buffer Additive, 0.3 μl Alexa488 and 127.5 μl) for 30 min at 20 °C (RT), and then rinsed with 3 % BSA. For BrdU staining, cells were rinsed twice with PBS, denatured using 2.5 M HCl (Fisher Scientific UK, Loughborough, Leicestershire, UK) for 1 h at RT, rinsed three times with PBS and two times with PBS+ (1 % BSA, 0.1 % Tween 20 (Sigma-Aldrich Company)). Cells were blocked with PBS+ for 1 h at 4°C before adding the primary rat (mESC; Abcam, Cambridge, UK) or mouse (hFb; Sigma-Aldrich Company) anti-BrdU antibody (diluted 1:1,000 with PBS+) for 2 h at RT. After rinsing the cells three times with PBS and three times with PBS+ the secondary Cy3-conjugated donkey anti-rat (mESC; Jackson Immuno Research Laboratories, West Grove, PA), Cy3-conjugated donkey anti-mouse (hFb; Jackson Immuno Research Laboratories), Alexa488-conjugated donkey anti-rat IgG (mESC; Molecular Probes Invitrogen) or Alexa-488-conjugated goat anti-mouse IgG (hFb; Molecular Probes Invitrogen) antibody (all diluted 1:1,000 with PBS+) was added for 40 min at RT in the dark. Cells were then washed three times with PBS and two times with PBS+ .
For DDR staining, cells were rinsed twice with PBS, twice with PBS+ and blocked in PBS+ for 1 h at 4 °C. As primary antibody either rabbit anti-γH2AX, rat anti-RPA32 or rabbit anti-53BP1 (Cell Signalling Technology, Danvers, MA; 1:500 dilution with PBS+) was added for 1 h, followed by rinsing three times with PBS (0, 5 and 10 min) and three times with PBS+ (0, 5 and 10 min). As secondary antibody either Cy3-conjugated donkey anti-rabbit or anti-rat (Jackson Immuno Research Laboratories; 1:500 dilution with PBS+) was added for 30 min at RT in the dark, followed by rinsing three times with PBS and two times with PBS+.
Samples were mounted in vectashield containing diamidinophenylindole (DAPI; Vector Laboratories, Peterborough, UK). Microscopy of the fixed and stained samples was performed using the Plan-APOCHROMAT 100×/1.40 Oil DIC objective from Zeiss on the Zeiss Axiovert 200 M Inverted Epifluorescence Microscope (Cambridge, UK). Images were obtained using a charge coupled device camera (International Power Sources, Holliston, MA) and Capture-Scope Software version 7.6.3.0 (MAG Biosystems). We note that while this style of wide-field microscope delivers images that have slightly inferior quality to confocal images because greater depth of field is imaged this does not impact on our ability to accurately score different patterns of incorporation. This is true even for ES cells, despite the challenge of delivering high quality wide-field images of these cells that grow in dense 3D colonies.
DNA fibres
HeLa cells were pulse labelled using 10 μM of either BrdU or EdU. DNA fibres were spread after 20 min and 2 h of incubation as is described by Jackson and Pombo (1998). BrdU detection was performed by incubation over-night with an anti-BrdU antibody (Rat anti-BrdU AbD Serotec, dilution 1:1,000) and a secondary detection using AlexaFluor-488 goat anti-rat (1:1,000 for 1 h). EdU-labelled fibres were detected using click chemistry with AlexaFluor-488. To enhance the signal of the click chemistry, samples were subsequently treated with anti-rabbit anti-AlexaFluor-488 (Life Technologies) followed by donkey anti-rabbit antibody conjugated with AlexaFluor-488 (Life Technologies), using conditions described above. After detection, slides were mounted in Prolong gold mounting media (Life technologies). LSM510 microscope (Zeiss) was used to acquire pictures of DNA fibres and the length of the fork measured using the LSM browser software.
Flow cytometry
Exponentially growing cells were plated on 6-well plates (mESC) or 60-mm dishes (hFb) (Corning), were labelled with 10 μM BrdU or EdU for 24 h, and either trypsinised immediately after labelling, or rinsed twice with medium before growing in fresh medium for an additional 24 h. Cell suspensions were washed with PBS and centrifuged for 5 min at 200×g. For DNA content analysis, cells were fixed in 50 % ethanol for at least 24 h at 4 °C, centrifuged for 10 min at 200×g, washed in PBS and spun down again (10 min at 200×g). Cells were resuspended in propidium iodide (PI) buffer (0.4 mg/ml RNase A (Sigma-Aldrich Company), 0.1 % Triton X-100 and 5 μg/ml PI (Gibco Invitrogen) in PBS and incubated for 30–45 min at RT. Fluorescence intensity was determined by flow cytometry on a Fluorescence Activated Cell Sorter Calibur (BD Biosciences, Oxford, UK) equipped with a 488-nm argon laser. Data acquisition was performed with the CellQuest (BD Biosiciences) software, and the percentages of G1-, S- and G2/M-phase cells were calculated with the Flowjo software program (Tree Star, Ashland, OR).
Cell counting
Exponentially growing cells plated on 6-well plates (Corning) were labelled with 10 μM BrdU or EdU for either 30 min or a time equivalent to the average cell cycle time for the cells used (15 h for mESC; 24 h for hFb, MEF and HeLa cells), rinsed twice with warm medium and chased in fresh medium for up to 48 h post-labelling. For counting on the automated cell counter (Bio-Rad Laboratories, Hemel Hempstead, Hertfordshire, UK), cells were trypsinised and stained with trypan blue solution (1:1 dilution; Sigma-Aldrich Company). Unlabelled cells were used as controls.
Results
Cell cycle progression in single cells
Replication precursor analogues such as BrdU and EdU have been used extensively in pulse-chase-pulse experiments to monitor S-phase progression. As this requires that S-phase is not perturbed by the incorporation of unnatural bases we evaluated if integration of EdU into the replicating DNA has any immediate effect on S-phase progression. Cells were pulse labelled with either a first pulse of BrdU or EdU (30 min) followed by a chase period of 4 h and then a second pulse using Cy3-dUTPs (30 min). Using fluorescent microscopy, the pulses showed non-overlapping signal with different replication patterns observed (Fig. 1a). Furthermore, counting the cells with one or two labels revealed no difference in EdU- and BrdU-labelled cells indicating that cells appear to progress through S-phase with similar rates under the different labelling conditions used (Fig. 1b). As the rate of S-phase progression will be directly dependent on the rate of fork elongation, we also measured for rate in BrdU- and EdU-treated cells (Fig. 2). HeLa cells were chosen for this analysis based on our extensive experience of the distribution of replication fork rates in this cell type (Jackson and Pombo 1998). HeLa cells were pulse labelled for 20 min or 2 h using either BrdU or EdU. DNA fibres were spread on glass slides (Fig. 2a), immuno-labelled and rates of fork progression calculated by measuring the length of replicated DNA stretches. Accurate estimates of the average fork rates were calculated during the 20 min pulse, during which time only a minority of forks will be completed (Fig. 2b). In the experiment shown, the average rate of fork migration in EdU-treated cells was indistinguishable from the rate seen in cells grown in medium supplemented with BrdU (Fig. 2; p value of 0.7793; n = 100). The extent of labelling on DNA spreads was also indistinguishable for BrdU and EdU after labelling for 2 h. However, after this longer period of incorporation individual fibres are generally much longer because of fusion of adjacent replicons so that reliable data on replication fork rates cannot be extracted (Fig. 2a). Even so, simple visual analysis of the spreads confirms that no gross disruption had occurred.
Cell cycle progression following EdU incorporation. Mouse embryonic stem cells (mESC) and human fibroblasts (hFb; MRC5) were labelled during replication with either BrdU or EdU for 30 min. After a chase time of 4 h cells were transfected with Cy3-dUTPs and incubated for 30 min. BrdU was detected using Alexa488-conjugated secondary antibody and EdU coupled directly with Alexa488. When stained cells were analysed using epi-fluorescence microscopy (a), the proportion of single- and double-labelled cells (b; n = 100) showed that S-phase progression was unaffected by EdU incorporation. Unperturbed S-phase progression was also evident from the distribution of early, mid-, and late replication patterns in double-labelled cells (a, b), and no differences were seen when EdU or BrdU was used. Scale bars, 5 μm
Replication fork speed is not affected by pulse labelling with EdU. HeLa cells were pulse labelled for 20 min or 2 h using 10 μM of either BrdU or EdU. Spread DNA fibres were visualised using indirect immuno-labelling with Aleax488 (a) and the rate of replication fork progression estimated from the length of the labelled regions (b). After labelling for 20 min, the mean fork rate in the presence of BrdU or EdU was 1.43 kb/min ± 0.38 (n = 100) and 1.41 kb/min ± 0.42 (n = 100), respectively. Scale bars, 10 μm
After cell division, stem cells might have the potential to keep one parental DNA strand dividing asymmetrically during cell differentiation (Lew et al. 2008). By analysing chromosome territories (CTs) in DNA-labelled cells we wanted to follow the fate of stem cells. Cells were labelled with EdU or BrdU for 30 min and incubated in fresh media for 3–6 days. With normal cell cycle progression it was expected to see one to three stained CTs under the microscope; the cells used have a cell cycle time of 15 h and as labelled CTs are randomly segregated during mitosis the labelled cells should contain only one to two CTs after seven division cycles. Indeed, in BrdU-labelled cells, only small numbers of isolated CTs could be detected (Fig. 3, top). However, after EdU labelling in mESC a dramatic loss of labelled cells was seen (data not shown); rare cells that survived apoptosis had aberrant nuclear morphology, typified by the abnormal nuclear shape shown (Fig. 3, EdU-labelled mESC). These surviving cells also retained uniform EdU labelling, which must result from activation of a robust cell cycle arrest and subsequent inhibition of proliferation. In contrast, EdU-labelled hFb displayed discrete labelled CTs (Fig. 3, HFb). However, the number of stained CTs/cell was higher than in the BrdU-treated controls, implying that their cell cycle timing is perturbed.
EdU inhibits cell cycle progression in mESC. mESC and MRC5 (hFb) cells were labelled during replication with either BrdU or EdU for 30 min and grown for a further five to six cell cycles in fresh medium. Incorporated BrdU and EdU was labelled as before (Fig. 1) and DNA counterstained with DAPI (blue). When cells were analysed using epi-fluorescence microscopy both mESC and hFb cultures labelled with BrdU showed the expected distribution of cells with isolated CTs (typical images shown in the first and third rows). However, after labelling with EdU, cells in mESC cultures had fully labelled nuclei with no cells displaying isolated territories (mESC/EdU) whereas MRC5 cultures had cells with isolated CTs but with more labelled sites than in BrdU-labelled cultures (hFb/EdU), consistent with delayed cell cycle progression in these cells. Bar, 5 μm
Cell cycle progression in cell populations
As variable defects in cell cycle progression were suggested in EdU-labelled cells, we next analysed the cell cycle profile in cell populations that were labelled throughout S-phase. Cells were labelled with EdU or BrdU for 24 h, stained with PI after fixation and then analysed using flow cytometry (Fig. 4; Table 1). EdU-treated mESC cell populations were clearly accumulated in G2/M (+47 %) with a corresponding decline of G1 cells (Fig. 4a). A shift to G2/M could also be detected for hFb (+24 %), however, the G1 population showed no difference to the control (Fig. 4b). Additionally, cells cultured in medium for 24 h post-labelling showed some minor accumulation in S-phase (+ 13 %) with a corresponding decrease in G1 cells (Fig. 4). BrdU-labelled cells treated in parallel showed small but significant difference in their cell cycle profile in comparison with the control. BrdU-treated mESC and hFb populations, directly fixed post-labelling, were shifted to G1 (+42 % in mESC and +15 % in hFb) (Fig. 4a, b), whereas, hFb-fixed 24-h post-labelling were shifted to S (+12 %) similar to EdU-treated hFb 24-h post-labelling (Fig. 4b). The results revealed that mESC cells labelled with EdU arrest cell cycle at either the G2/M or M checkpoint. Conversely, hFb show only a weak response to EdU treatment. In conclusion, EdU incorporation results in cell cycle arrest in mESC after S-phase completion. hFb in contrast appear to escape significant cell cycle arrest, though their progression through the cell cycle is delayed.
EdU-labelled mESC cultures arrest cell cycle in G2/M. mESC and MRC5 (hFb) cells were treated with BrdU or EdU for 24 h and either fixed immediately or washed and incubated for additional 24 h in fresh medium. DNA was stained with PI and cell cycle distributions analysed by flow cytometry, using untreated cultures as controls. For mESC cultures (a), incorporation of BrdU resulted in an increase in G1/S cells and clear reduction of cells in G2/M whereas incorporation of EdU had the opposite effect with a dramatic increase in G2/M cells and many fewer cell in G1/S. For hFb cultures (b), BrdU incorporation had no significant effect on cell cycle progression while incorporation of EdU increased the number of cells in G2/M, consistent will retarded cell cycle progression in these cells. For each sample, typical flow profiles (20,000 cells/sample) are shown (left) and the cell cycle distribution of cells in G1, S, and G2/M shown (right; error bars show standard deviation of the mean; n = 3)
In order to test the effect of EdU on cell viability, we labelled different cells types using BrdU or EdU as before. Short pulses of the nucleotide analogue (30 min) or continuous labelling (15–24 h depending on the cell cycle length in the particular cell type) were added to the media and the number of cells counted at 24-h intervals for up to 3 days. For short pulses, because unsynchronised cell populations will be spread uniformly across the cell cycle, some cells will be labelled (S-phase cells during the EdU pulse) and some will be unlabelled (non-S-phase cells during the EdU pulse) after incubation in EdU. Even so, analysis of cell viability showed that EdU-treated cells have compromised cell viability, consistent with EdU incorporation either delaying the progression of cells through the cycle and/or inducing cell death (Fig. 5a; Table 2). The conclusion was confirmed when cells were grown in the presence of EdU for an entire cell cycle. In this case, all cell types studied showed retarded growth characteristics with significantly lower cell numbers than untreated control samples at all time points analysed (Fig. 5b). However, mESC showed the most profound proliferation defects after EdU treatment, with an almost complete block of cell cycle progression, and hence proliferation.
Prolonged incorporation of EdU induces cell death. mESC, MRC5 (hFb), mouse embryonic fibroblasts (MEF), and human cancer cells (HeLa) were labelled with BrdU or EdU during short (a; 30 min) or long (b; approximately 1 cell cycle; pulse times shown above individual histograms) pulses, washed, and incubated in fresh medium for up to 48 h. Cell proliferation was monitored by automated cell counting and trypan blue staining used to identify dying cells. Untreated cells were used as controls. Histograms show arithmetic means of the relative cell numbers (n = 3; error bars show standard deviations) with cell numbers at the onset of labelling set to 100 %
DNA damage response
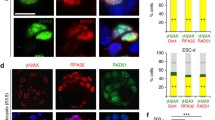
As EdU incorporation clearly impacts on the efficiency of cell cycle progression we next analysed if this was a consequence of inducing a robust DDR in the EdU-treated cells. Following brief periods of culture in EdU, mESC were processed for immuno-histochemistry and major proteins involved in the DDR—γH2AX, RPA32 and 53BP1—visualised by indirect immune-labelling to monitor the extent of DDR induction (Fig. 6). In this series of experiments, cells were pulse labelled with EdU or BrdU for 30 min (incorporation index is ∼70 % for mESCs and ∼30 % for hFbs) or 24 h (∼100 % mESCs and hFbs labelled) and then grown for 24 h before analysis in order to assess levels of expression of the DDR proteins.
EdU triggers DDR in mESC. mESC cells were treated with BrdU or EdU for 30 min, washed, and incubated for 24 h. DDR was detected with anti-γH2AX, anti-RPA32 or anti-53BP1 primary antibodies and Cy3 (red) conjugated secondary antibody. The nucleus was counterstained using DAPI (blue). Cells were analysed using epi-fluorescence microscopy (a) and cells with a positive DDR assessed by comparison with unlabelled control cells (b; n = 150 cells in two independent experiments). Based on control cultures, cells with <5 (RPA32) or <10 (γH2AX, 53BP1) sites were scored “negative” and cells with more spots were scored “positive”. Bar, 5 μm. (**P < 0.01; ***P < 0.001; ****P < 0.0001)
As some basal level of DDR induction is known to exist in the absence of exogenous damage, we monitored characteristic DDR foci in untreated mESC to establish maximum levels of background expression. For γH2AX and 53BP1, in different experiments, ∼10 % of cells were positive and for RPA32 ∼12 %; hence, 10 % of positive cells was set as the background levels for γH2AX/53BP1 and RPA32 (Fig. 6). In EdU-treated mESC, significant increases in DDR-positive cells were seen when compared with untreated controls (Fig. 6), with changes in nuclear 53BP1 showing the most profound effects, both in terms of positive cells and the number and intensity of nuclear foci following EdU incorporation. Widespread and persistent formation of γH2AX foci was also seen in these cells (Fig. 6). In comparison, BrdU incorporation correlated with only a slight increase in the formation of γH2AX and no significant change in the formation of RPA32 and 53BP1 foci (Fig. 6). Hence, incorporation of EdU into mESC DNA clearly activates a robust DDR, which is very different in magnitude to the slight effect seen following BrdU incorporation for the same amount of time.
Similar trends were seen in hFb with robust activation of DDR in about 50 % of cells following EdU incorporation and much less significant induction of DDR in cultures treated with BrdU (Fig. 7). Though qualitatively similar responses are seen in these two cell types it is important to emphasise quantitative differences. Notably, the mESC data shown in Fig. 6 resulted from a short period of EdU incorporation (only 30 min), during which time ∼70 % of cells incorporate EdU into their DNA. Essentially, all EdU-containing cells activate some features of DDR. In contrast, hFbs with similar levels of DDR induction contain much higher levels of EdU, which in the experiment shown in Fig. 7 is incorporated throughout their S-phase. In this case, while all cells in the culture will have incorporated EdU only ∼50 % activate robust DDR. Hence, the extent of EdU-induced DDR activation in these different cell types correlates with their ability to either arrest and die by apoptosis (mESC) or progress into subsequent cell proliferation cycles (hFb).
EdU triggers DDR in hFb. MRC5 cells were treated with BrdU or EdU for 24 h, washed and incubated for an additional 24 h and the DDR assessed as described in the legend to Fig. 6. Bar, 5 μm (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001)
Discussion
Many different replication precursor analogues have been used over the years to monitor S-phase progression and the distribution of replicated DNA into subsequent generations. Largely, because of ease of use and reliability of labelling following incorporation, the commercially available compound EdU has been adopted over the past 3–4 years as the reagent of choice for labelling replicated DNA. However, despite the routine use of EdU and demonstration that in some cell types EdU incorporation appears not to affect cell cycle progression (Aparicio et al. 2012) in other studies EdU is seen to inhibit cell proliferation (Meneni et al. 2007; Diermeier-Daucher et al. 2009; Ross et al. 2011). This inhibitory effect of EdU on cell cycle progression suggests that the cytotoxic consequences of high doses of EdU might even be used for cancer therapy (Diermeier-Daucher et al. 2009; Ross et al. 2011). As EdU incorporation appears to have variable cell type and concentration specific effects of cell proliferation, despite its routine use for labelling DNA, it is essential to evaluate the extent of any cytotoxic effects. With this in mind, we have evaluated how EdU influences cell proliferation using a range of cell types - mESC, hFb, MEF and HeLa - which to date have not been analysed for EdU-dependent proliferation phenotypes. We show that EdU does indeed compromise the rate of cell proliferation in different cell types and that profound differences in the EdU-dependent responses are seen.
We used EdU both for pulse labelling and continuous labelling, throughout S-phase, at concentrations recommended by the supplier and found that the incorporation of EdU had little or no effect on the structure of the S-phase programme (Fig. 1) or the rate of fork progression (Fig. 2) when applied for short times. However, when flow cytometry was used to monitor cell cycle progression a clear disruption of the cell cycle could be observed, with cells accumulating in G2/M (Fig. 3). The extent of accumulation was variable, depending on the cell type used. Careful analysis of cell numbers showed that this cell cycle perturbation also correlated with a decline in cell number (Fig. 5), with some cells being retarded in their cell cycle progression (Fig. 3) and others dying (Figs. 3 and 5b). This correlated in turn with induction of a robust DDR, with the DDR activation having an impact on the decline in proliferative potential (Figs. 6 and 7). Notably, DDR induced by EdU was much more profound than that activated by BrdU incorporation. From structural similarities, it is reasonable to propose that the reactive alkyne group on the EdU molecule has a more potent ability to damage DNA than the bromo group, which under some conditions is known to induce low levels of DNA damage and sister chromatid exchange (Meneni et al. 2007). However, it was clear in our studies that the EdU-induced DDR was quite variable between cell types, with low levels of EdU incorporation giving a very robust and persistent response in mESC, which typically resulted in cell death, whereas hFb were able to re-enter cell cycle following DDR-induced arrest when much higher levels of EdU incorporation were used.
Taking these results together, it can be concluded that the use of EdU for DNA labelling is limited, as some degree of EdU-dependent cell cycle perturbation and cell death will occur in prolonged culture. As the extent of cell death in different cell lines correlates with the amount of EdU incorporated, minimising the concentrations of EdU used during labelling is likely to limit its impact on cell proliferation (Diermeier-Daucher et al. 2009; Ross et al. 2011). Perhaps the most notable feature of cellular response to EdU labelling is the substantial variability in response by different cell types, as discussed above. Clearly, this variability in the way in which cells respond to EdU incorporation demonstrates that great care must be taken in assessing cell proliferation when EdU is used to analyse different aspects of DNA replication and chromosome organisation in eukaryotic cells.
Abbreviations
- BrdU:
-
Bromodeoxyuridine
- CT:
-
Chromosome territory
- DAPI:
-
Diamidinophenylindole
- DDR:
-
DNA damage response
- EdU:
-
Ethynyldeoxyuridine
- hFb:
-
Human fibroblasts
- mESC:
-
Mouse embryonic stem cells
- PI:
-
Propidium iodide
References
Aparicio T, Megías D, Méndez J (2012) Visualization of the MCM DNA helicase at replication factories before the onset of DNA synthesis. Chromosoma 121:499–507
Blow JJ, Gillespie PJ, Francis D, Jackson DA (2001) Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol 152:15–25
Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A (2008) Detection of S-phase cell cycle progression using 5-ethynyl-2-deoxyuridine incorporation with click chemistry an alternative to using 5-bromo-2-deoxyuridine antibodies. Biotechniques 44:927–929
Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G (2009) Cell type specific applicability of 5-ethynyl-2-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A 75A:535–546
Edenberg HJ (1976) Inhibition of DNA replication by ultraviolet light. Biophys J 16:849–860
Fakan S, Turner GN, Pagano JS, Hancock R (1972) Sites of replication of chromosomal DNA in a eukaryotic cell. Proc Natl Acad Sci U S A 69:2300–2305
Farkash-Amar S, Simon I (2010) Genome-wide analysis of the replication program in mammals. Chromosome Res 18:115–125
Jackson DA, Pombo A (1998) Replicon cluster are stable units of chromosome structure: evidence that nuclear organisation contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140:1285–1295
Lew DJ, Burke DJ, Dutta A (2008) The immortal strand hypothesis: how could it work? Cell 133:21–23
Maya-Mendoza A, Tang CW, Pombo A, Jackson DA (2009) Mechanisms regulating S phase progression in mammalian cells. Front Biosci 14:4199–4213
Maya-Mendoza A, Olivares-Chauvet P, Shaw A, Jackson DA (2010) S phase progression in human cells is dictated by the genetic continuity of DNA foci. PLoS Genet 6:e1000900
Meneni S, Ott I, Sergeant CD, Sniady A, Gust R, Dembinski R (2007) 5-alkynyl-2-deoxyuridines: chromatography-free synthesis and cytotoxicity evaluation against human breast cancer cells. Bioorg Med Chem 15:3082–3088
Nakamura H, Morita O, Sato C (1986) Structural organisations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res 165:291–297
Olivares-Chauvet P, Fennessy D, Jackson DA, Maya-Mendoza A (2011) Innate structure of DNA foci restricts the mixing of DNA from different chromosome territories. PLoS One 6:e27527
Petruk S, Sedkov Y, Johnston DM et al (2012) TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150:922–933
Philimonenko AA, Hodný Z, Jackson DA, Hozák P (2006) The microarchitecture of DNA replication domains. Histochem Cell Biol 125:103–117
Ross HH, Rahman M, Levkoff LH et al (2011) Ethynyldeoxyuridine (EdU) supresses in vitro population expansion and in vivo tumor progression of human glioblastoma cells. J Neurooncol 105:485–498
Salic A, Mitchison TJ (2008) A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105:2415–2420
Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer TJ (2003) Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. Cell Biol 160:685–697
Acknowledgments
We thank the BBSRC for funding that contributed to the work described here. We also thank Werner Mueller (Manchester, UK) for providing the ES cell line and Ricardo Rodriguez-Enriquez (Manchester, UK) for providing the MEF cell line.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Wendy Bickmore.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Kohlmeier, F., Maya-Mendoza, A. & Jackson, D.A. EdU induces DNA damage response and cell death in mESC in culture. Chromosome Res 21, 87–100 (2013). https://doi.org/10.1007/s10577-013-9340-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-013-9340-5