Abstract
Chromosome architecture and assorted nuclear compartments play an essential role in RNA transcription and processing. Growing vertebrate oocytes represent an advantageous model to study the principles of nuclear structure and function. In this article, the data on three-dimensional (3D) organisation of intact and non-deformed oocyte nuclei (germinal vesicles) in four species of birds (domestic chicken, Japanese quail, rock pigeon and chaffinch) obtained by confocal laser scanning microscopy are presented. The nucleus of the growing avian oocyte has an unusual structure of RNA processing machinery. Germinal vesicles from any of the avian species studied, with the exception of the rock pigeon, are characterized by absence of extrachromosomal RNA-enriched nuclear bodies including Cajal bodies, histone locus bodies and interchromatin granule clusters. The absence of Cajal bodies and histone locus bodies in chicken oocytes correlated with the inactivation of nucleolus organizer and clustered histone genes. Splicing factors such as SR-protein SC35 accumulated in chromosome-associated domains that were classified as complex loops (terminal giant and lumpy loops). Formation of such depot in avian oocyte nuclei is supposed to be nucleated by transcripts of non-coding tandem repeats. The results obtained strongly support a model of RNA-mediated nuclear domains formation.
Similar content being viewed by others
Introduction
The structural compartmentalization of the cell nucleus, including the spatial chromatin arrangement and formation of assorted nuclear organelles, that concentrate various components of RNA splicing, 3′-processing and export machineries, play an essential role in the regulation of eukaryotic gene expression (Kumaran et al. 2008). The main principle behind nuclear organisation is to segregate the intranuclear space into a number of highly dynamic but functionally stable compartments and subdomains related to individual nuclear processes (Kumaran et al. 2008; Morris 2008; Tripathi and Prasanth 2011). These domains include the most prominent and evolutionarily conserved nuclear compartments such as nucleoli, splicing-factor domains and Cajal bodies (CBs), as well as smaller domains such as replication foci and transcription factories. A functional link between the formation of various nuclear organelles and the activity of defined genomic sequences is well established in the case of such compartments as the nucleolus and histone locus body (HLB) (Tripathi and Prasanth 2011). For instance, HLBs form in association with transcribed clusters of histone genes and store components involved in the specific 3′ processing of histone pre-mRNAs such as U7 small nuclear RNA (snRNA), stem loop binding protein and symplekin (Nizami et al. 2010; Shevtsov and Dundr 2011).
The exploration of the functional relations between nuclear organelles and genomic loci requires manipulation of gene expression (Lemm et al. 2006). An alternative and perspective approach may be to investigate the structural and functional variations in nuclear structure that exist in nature (Liu et al. 2006; Bogolyubov and Parfenov 2008).
The nucleus of a growing amphibian oocyte provides an advantageous experimental system to study the functional compartmentalization of cell nucleus (Morgan 2002; Gall et al. 2004; Morgan 2008; Gall and Wu 2010; Sommerville 2010). Likewise, an avian oocyte nucleus could also be employed as a model to explore the three-dimensional (3D) organisation of chromosomes and other nuclear structures (Maslova and Krasikova 2011; Khodyuchenko et al. 2012; Maslova and Krasikova 2012). The oocyte nucleus was first described by Jan Evangelista Purkyně who examined transparent structure inside chicken oocytes in the early 1830s and named this organelle ‘vesicula germinativa’, or the germinal vesicle (GV) (De La Fuente 2006). The main advantage of this system is the giant size of the nucleus, which can be up to 400 μm in diameter. Another benefit is that the chromosomes in the oocyte nucleus of these animals acquire a characteristic ‘lampbrush’ form (reviewed in Callan 1986; Morgan 2002; Gaginskaya et al. 2009). Within each lampbrush meiotic bivalent, homologous chromosomes are represented by an array of chromomeres, with pairs of long transcriptionally active loops extending laterally. Importantly, most of the individual transcription units in lampbrush chromosomes (LBCs) can be visualized by light microscopy, and co-transcriptional stages of RNA processing can be studied in detail (reviewed in Morgan 2002; Gaginskaya et al. 2009). Moreover, cytological maps indicating the relative positions of major landmark structures such as associated nuclear bodies and complex loops were created for many amphibian species (Callan and Lloyd 1960; Callan 1986; Macgregor et al. 1990). For certain avian species such as chicken and Japanese quail, detailed high-resolution cytological chromomere-loop maps of lampbrush chromosomes with a number of genomic fragments assigned to either chromomere or lateral loops are currently also available (Galkina et al. 2006; Zlotina et al. 2012).
Due to the tight cooperation between the oocyte and follicle cells, oocyte–follicle cell complexes can be employed as a potential model to investigate the variations in nuclear body formation as determined by the status of the oocyte genome. In animals in which the oocyte passes through a transcriptionally active stage, the transformation of chromosomes into the lampbrush form could be unaccompanied by the activation of housekeeping genes within the oocyte nucleus. For instance, in adult birds from different orders, the inactivation of the nucleolus organizer in growing oocytes is demonstrated at both the cytological and cytogenetic levels (Greenfield 1966; Gaginskaia and Chin 1980; Kropotova and Gaginskaya 1984; Hutchison 1987). In contrast with the oocytes of many other vertebrates, such as amphibians and some reptiles, avian large-sized oocytes from egg-laying females do not house any amplified nucleoli (Gaginskaia 1972; Gaginskaia and Gruzova 1975; reviewed in Gaginskaya et al. 2009). Nucleoli appear only in small growing oocytes (less than 200 μm in diameter) of 1-month-old and younger avian females (Greenfield 1966; Chin et al. 1979).
Nuclear compartmentalization and spatial genome configuration in growing vertebrate oocytes has been most widely studied in amphibian, mammalian and insect oocytes (Gall et al. 2004; De La Fuente 2006; Bogolyubov and Parfenov 2008; Morgan 2008), whereas the 3D-structure of avian GVs is less investigated. The morpho-functional organisation of RNA splicing and processing machineries apart from the absence of functional nucleolus in transcriptionally active avian oocytes remained largely unclear.
In this article, we describe the 3D organisation of sub-nuclear structures such as lampbrush chromosomes and nuclear domains in oocytes from four avian species (domestic chicken, Japanese quail, rock pigeon and chaffinch) using confocal laser scanning microscopy (CLSM) and computational image processing procedures (iso-surfacing and 3D-reconstruction). These approaches provide additional clues to the functional compartmentalization of the avian oocyte nucleus.
Materials and methods
Materials and objects
Oocytes of the domestic chicken (Gallus gallus domesticus), Japanese quail (Coturnix coturnix japonica), rock pigeon (Columba livia), chaffinch (Fringilla coelebs), African clawed frog (Xenopus laevis), lake frog (Pelophylax ridibundus) and pool frog (Pelophylax lessonae) were used in this study. All manipulations with animals were carried out in accordance with the “Committee for the Update of the Guide for the Care and Use of Laboratory Animals (2011)”. Prior to ovary isolation, frogs were anaesthetized with MS222 (tricaine methanesulfonate) at 1.5 g/L (Sigma); a small part of the ovary was surgically removed from the frog. Oocytes were incubated at 18 °C in OR2 medium (82.5 mM NaCl, 2.5 mM KCl, 1.0 mM CaCl2, 1.0 mM MgCl2, 1.0 mM Na2HPO4, 5.0 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid)) up to 24 h. Ovaries, obtained from chicken, quail, pigeon or chaffinch, were placed into a small weigh bottle with a piece of filter paper, moistened with “5:1 + phosphates” medium (83.0 мМ KCl, 17.0 mM NaCl, 6.5 mM Na2HPO4, 3.5 mM KH2PO4, 1 mM MgCl2, 1 mM dithiothreitol) and were kept at 4 °C for not more than 1 h.
Oocyte nucleus isolation and fixation
Amphibian and avian oocyte nuclei were isolated in the “5:1 + phosphates” medium from oocytes of 0.5–1 mm and 0.7–20 mm in diameter, respectively, according to the standard procedures (http://projects.exeter.ac.uk/lampbrush/protocols.htm). Oocytes after removal from the ovary were kept in “5:1+ phosphates” medium at 4 °C for not more then 5 min. Small holes in the oocyte envelope and membrane were made with thin tungsten needles in the avian oocytes and by fine jeweler forceps in the amphibian oocytes. Nuclei were carefully squeezed out and cleaned of yolk granules and ooplasm. Oocytes and nuclei were inspected with a Leica MZ16 stereomicroscope during all manipulations. For avian germinal vesicles, only nuclei from the healthiest oocytes (partially transparent and not whitish) were used. Avian germinal vesicles were fixed in 2 % paraformaldehyde (PFA) for 30 min at room temperature immediately after isolation and then washed in phosphate-buffered saline (PBS, 1.47 mМ KH2PO4, 4.29 mM Na2HPO4 × 7H2O, 137 mM NaCl, 2.68 mM KCl, pH 7.3). Isolated nuclei were transferred from one embryo cup to another with a pipette. It was essential to prevent nuclei from sticking to the glass bottom of the embryo cup or liquid surface by putting the nucleus at the bottom of the embryo cup and moving the cup carefully to generate medium flow. Fixed avian GVs could be stored in PBS in humidified chambers at +4 °C for at least 4 days.
Staining with fluorochromes
To investigate the content of intact oocyte nuclei by means of confocal microscopy, isolated nuclei were placed in chambers filled with “5:1 + phosphates” medium with 0.07 μМ nucleic acid-specific dye Sytox green (Molecular Probes). The microscopy chambers were constructed by sticking a plastic square with a hole to a cover glass 24 × 50 × 0.16–0.19 mm (Deckgläser, Menzel-glaser) with paraffin and Vaseline mixture (1:1). A small excess of the medium was required in the observation chamber (convex meniscus) to prevent bubbles from forming during the covering of the chamber by a cover glass. After mounting, the 3D-preserved nuclei were immediately examined by confocal laser scanning microscopy, to prevent overstaining the GV with fluorochromes.
Immunofluorescent staining of oocytes and isolated oocyte nuclei
Prior to immunofluorescent staining, dissected avian oocytes were treated with 1 mg/ml collagenase (Sigma) in PBS for 30 min to improve antibody (Ab) penetration. After a series of washes, the oocytes were fixed in 2 % PFA (Serva), permeabilized in 0.2–0.5 % Triton X-100 (ICN Biomedicals), blocked with 1 % blocking reagent (Roche) and incubated with primary and secondary Abs as described previously (Khodyuchenko et al. 2012). Oocytes were mounted in antifade DAВCO mounting medium (Мerck), containing 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/ml; Sigma).
Immunofluorescent staining of fixed nuclei was performed as described previously (Khodyuchenko et al. 2012). Nuclei were incubated for 1 h with primary Abs diluted in PBS, washed with PBS for 10 min and incubated with the appropriate secondary Abs diluted in PBS for 1 h. Commercial blocking solutions were not applicable for GV incubation due to the substantial nuclear shrinkage and loss of 3D morphology. After the final washes, immunostained GVs were transferred into glass chambers and analysed with confocal laser scanning microscope.
Antibodies used
The following primary Abs were used: rabbit pAb H-300 against coilin (dilution 1:100; Santa Cruz Biotechnology); mouse monoclonal antibody (mAb) against serine/arginine-rich protein (SR-protein) splicing component (factor) 35 kDa (SC35, dilution 1:50; Abcam); mouse mAb Y12 against symmetrical dimethylarginine (dilution 1:50) (Lerner et al. 1981); mouse mAb K121 against 2,2,7-trimethyl guanosine (dilution 1:150; Santa Cruz Biotechnology) and mouse mAb 3С2 against heterogeneous nuclear ribonucleoprotein (hnRNP) K/J (dilution 1:500) (Matunis et al. 1992). The following secondary Abs were used: Cy3-conjugated goat anti-rabbit IgG (dilution 1:500) and Alexa-488-conjugated goat anti-mouse IgG (dilution 1:300; Jackson ImmunoResearch Laboratories).
3D-FISH
3D fluorescent in situ hybridization (FISH) on preserved GVs with large volumes was performed without denaturation of genomic DNA that allows the maintenance of the 3D architecture of the giant nucleus. Cy3- and biotin-labelled single stranded oligonucleotide probes (Syntol, Moscow) were used for FISH that allowed to reduce the hybridization temperature. Fluorochrome-labelled probes are strongly preferable over hapten-labelled probes because they do not require detection steps and reduce nuclear loss. Fixed nuclei were placed in hybridization buffer (900 mM NaCl, 20 mM Tris–HCl, 0.1 % sodium dodecyl sulphate, pH 7.2) (Chen et al. 1997) which was diluted 1:8 in PBS for 1 h at room temperature. The nuclei were then transferred into hybridization buffer containing 10–20 ng/μl of the oligonucleotide probe and were incubated for 1 h at 48 °С in the dark. The nuclei were then washed multiple times in 0.4× saline–sodium citrate buffer (3 M NaCl, 0.3 M sodium citrate, pH 7) at 48 °С for 5 min each, mounted in glass chambers, containing PBS and DAPI (1 μg/ml) and immediately analysed by CLSM.
Confocal laser scanning microscopy
Series of optical sections through 3D-preserved nuclei and oocytes were obtained using the Leica TCS (True Confocal Scanner) SP5 confocal laser-scanning microscope equipped with HCX PL APO lambda blue oil immersion objectives and diode (405 nm), argon and helium-neon (543 nm) lasers (Leica Microsystems, Germany). For each optical section, signals in different channels were acquired sequentially then merged using LAS AF (Leica Application Suite) software (Leica-Microsystems, Germany). To reduce noise in the acquired images, each section was frame-averaged three times. Computational analyses of the 3D-image stacks and reconstructions were performed using the “Process” (“Colors”, “Crop” and “3D Projection”) and “Quantify” (“Line Profile”) tabs in the LAS AF software.
To visualize clearly nuclear structures, 3D images were processed with the Imaris 7.5 (Bitplane, AG) and ImageJ software, as described in detail by Maslova and Krasikova (2011). To improve the image quality of nuclear 3D reconstruction, the “Gaussian Blur” and “Background Remover” automatic program filters were applied. Isosurfaces were constructed around voxels with adjusted fluorescence intensities within a 3D-rendered nuclear volume so that voxels from individual chromosomes were incorporated into separate objects. Geometrical characteristics of isosurface objects, for instance a centre of gravity, were automatically extracted and used for spatial measurement of chromosome distribution within the oocyte nuclei.
FISH on isolated lampbrush chromosomes
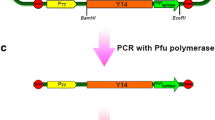
Avian lampbrush chromosome preparations were obtained by a standard technique (for references see Gaginskaya et al. 2009). Fluorescent in situ hybridization with bacterial artificial chromosome (BAC) clones on avian LBCs was performed as described previously (Galkina et al. 2006) according to DNA/RNA and DNA/(DNA + RNA) hybridization protocols. Biotin- and digoxigenin-labelled probes of BAC clones WAG115D22 (histone locus, 50 Mb, chromosome 1) and WAG053E23 (LEI0071, 79.4 Mb, chromosome 1) from the Wageningen chicken BAC library (Crooijmans et al. 2000) were used for hybridization. The following primers were used to verify the presence of chicken histone H2B gene in the ВАС clone WAG115D22 by standard polymerase chain reaction (PCR):
-
H2BF 5′-AAGGGCTCCAAGAAGGCGG-3′
-
H2BR 5′-GACGTGATGGTCGAGCGCTTG-3′
Probes for FISH were obtained by degenerate oligonucleotide primed PCR (DOP-PCR) DNA labelling, using BAC clone DNA as a template, 6 MW primer (Telenius et al. 1992) and either biotin-dUTP or digoxigenin-dUTP.
Biotin-labelled probes were detected with Alexa488-conjugated avidin, and enhanced by biotinylated goat antibodies against avidin; digoxigenin-labelled probes were detected with Cy3-conjugated anti-digoxigenin antibody (Jackson ImmunoResearch Laboratory). Slides were mounted in antifade DAВCO mounting medium (Мerck) containing DAPI (1 μg/ml). Preparations were examined under Leica DM 4000B microscope (Leica-Microsystems, Germany).
Results
Approach for three-dimensional analysis of avian oocyte nucleus topology
Avian GV spreads are not very suitable for studying the extrachromosomal nuclear bodies in avian oocytes. Even full sets of lampbrush chromosomes can quite rarely be found within a single spread (Hutchison 1987) and is rather an exception than a rule (approximately 0.5 % of spreads). In avian ovary, the number of healthy large growth stage oocytes of suitable sizes is limited and does not usually exceed 100. Moreover, the data on appearance, presence or absence of extrachromosomal non-nucleolar bodies in avian oocyte nuclei, based on observations of spread GV preparations, was controversial. In this study we examined the 3D organisation of oocyte nuclei from four species of birds (F. coelebs, C. livia, C. coturnix japonica and G. gallus domesticus) by confocal laser scanning microscopy. For that purpose, we introduced a variety of techniques that allowed us to analyse the 3D architecture of avian oocyte nuclei without sectioning or spreading.
It is known that GVs can be manually isolated from avian oocytes with either forceps or needles. Isolated intact and non-deformed (3D preserved) nuclei can be used for immediate observation or fixed and then be subjected to staining with fluorochromes, immunofluorescent staining or 3D-FISH (Fig. 1). After staining or FISH, the nucleus is transported to a chamber (well slide) in a phosphate buffer solution or nuclear isolation medium. The 3D-preserved oocyte nucleus lying on the bottom of the chamber can then be analysed directly by CLSM on the base of an inverted microscope (Fig. 1).
Technique for three-dimentional (3D) analysis of avian oocyte nucleus (germinal vesicle) architecture via confocal laser scanning microscopy. Manually isolated 3D-preserved oocyte nucleus can be fixed and then stained by fluorochromes or antibodies. 3D FISH according to DNA/RNA hybridization protocol can be applied to reveal the distribution of transcripts
Compared with studying the isolated GV content, the main advantage of the suggested approach is opportunity to analyse the 3D topology of the intranuclear structures in detail. Another advantage is a chance to visualize all chromosomes and extrachromosomal bodies in a single GV and to compare the concentration of RNA and proteins in different intranuclear structures and the nucleoplasm. On the other hand, there are some problems of working with 3D preserved GVs, including microsurgical isolation of the nucleus, skills required to handle the isolated nuclei and the low percentage of 3D preserved nuclei after indirect immunofluorescent staining or 3D FISH.
3D-organisation of lampbrush chromosomes in avian germinal vesicles
To analyse the general morphology of avian oocyte nuclei, undamaged GVs were stained with a NA-specific dye, Sytox green. As expected, a full set of lampbrush chromosomes that stained brightly with Sytox green was observed in the GVs of all avian species studied (Fig. 2). The overall 3D structure of the LBCs and nuclear bodies was not affected by nuclear fixation procedures (Video 1 and 2, Supplementary). Simple lateral loops (transcriptionally active loci) protruding from the brightly stained chromomeric axis (compact chromatin domains) of LBCs and complex loops can be clearly distinguished in fixed GVs (Fig. 2h, i; Video 1 and 2, Supplementary).
Diversity of nucleic acid-containing structures in growing avian and amphibian oocyte nuclei. a–e Maximum projection views of intact nuclei isolated from growing oocytes of the G. gallus domesticus (a), C. livia (b), F. coelebs (c), X. laevis (d) and P. ridibundus (e). Arrow points to extrachromosomal RNA-enriched spherical body in pigeon germinal vesicle. f Individual confocal section through chaffinch germinal vesicle. Centromere protein bodies (arrows) do not contain RNA. Related Supplementary materials—Video 2. g Individual lampbrush bivalent of clawed frog with histone locus bodies (arrows) attached. h, i Maximum projection of optical slices through fragments of the nucleus shown on (c) demonstrating individual chaffinch lampbrush chromosomes. Arrows point to simple lateral loops (h) and complex terminal giant loops (i). Staining with the nucleic acid specific fluorescent dye Sytox green. Scale bars a, b, e, f 50 μm, c 20 μm, d 100 μm, g–i 10 μm
Reconstructions and image segmentation of the scanned GVs from the Z-stacks of optical sections (Fig. 3a) can be used for lampbrush bivalents morphometrics, sorting and approximating of their positions (Maslova and Krasikova 2011). Isosurfaces corresponding to individual lampbrush bivalents can be computer-painted into different colours (Fig. 3b and Supplementary Video 3, Supplementary). In our previous study, using three quantitative approaches we demonstrated that there is no preference for gene-rich microchromosomes to be located at the nuclear interior in the chicken and quail GVs (Maslova and Krasikova 2011). Here we confirmed that at the lampbrush stage, gene-rich or gene-poor chromosomes demonstrate no preferential radial distribution by visualization of the positions of centres of gravity in the 3D GV reconstructions (Video 4, Supplementary).
Nuclear architecture in maturing avian oocytes at different stages of oocyte growth. a, b Projection view of isosurface-rendered optical stacks obtained by confocal laser scanning microscopy of isolated chicken (G. gallus domesticus) germinal vesicle stained with Sytox green. a Selected sex ZW bivalent isosurface is shown in yellow. b Thirty-nine bivalent isosurfaces are visualized in different colours. Related Supplementary materials—Video 3 and 4. c Maximum projection view of Japanese quail (C. coturnix japonica) germinal vesicle isolated from yellow-yolk late oocyte and stained with Sytox green; dashed line depicts nuclear border. Transcriptionally inert spherical chromosomes (green) are compacted into a karyosphere (arrow). d Maximum projection of optical slices through enlarged karyosphere. Scale bars: a–c 50 μm; d 5 μm
Nuclear domains structured by the non-coding transcripts of 41 bp tandem repeats
To elucidate the distribution of non-coding transcripts in 3D preserved avian oocyte nuclei we used the developed 3D FISH protocol and performed 3D FISH on Galliform germinal vesicles with probes for short 41 bp tandem repeats. Tandem PO41 (pattern of 41 repeat) and CNM (chicken erythrocyte nuclear membrane repeat) repeats in chicken or PO41 and BglII-repeats in Japanese quail were previously found to be intensively transcribed on a number of lampbrush chromosomes (Krasikova et al. 2006; Deryusheva et al. 2007). In both chicken and Japanese quail intact oocyte nuclei, we observed nuclear domains approximately 10 μm in size that contained nascent non-coding RNA of 41 bp tandem repeats (Fig. 4a). At stage of the most intensive transcription, these brightly labelled domains corresponded to polarized RNP-matrix of 3D-preseved satellite DNA transcription units up to 40 μm long (Fig. 4b–d). Based on these data and previous findings, we argue that in growing G. gallus domesticus and C. coturnix japonica oocytes, the intranuclear domains that are highly enriched with RNA of short 41 bp tandem repeats correspond only to the transcription units in the lampbrush chromosome lateral loops. Additionally, smaller and dimmer foci were radially distributed around the sites of satellite DNA transcription (Fig. 4c). These foci represented extrachromosomal complexes of RNP particles that comprise newly synthesized transcripts of tandem 41 bp repeats.
Nuclear domains enriched by transcripts of tandem repeats correspond to individual transcription units of lampbrush chromosomes. Maximum projection views of intact chicken (G. gallus domesticus) (a) and Japanese quail (C. coturnix japonica) (c) oocyte nuclei after three-dimensional FISH with a probe to PO41 tandem repeat (red) according to DNA/RNA hybridization protocol. b, d Maximum projections of optical slices through enlarged regions of the oocyte nuclei showing individual PO41 repeat transcriptions units. Scale bars a, c 50 μm; b, d 10 μm
In spread preparations of chicken and Japanese quail lampbrush chromosomes, the hnRNP K protein was found to be enriched in the RNP-matrix of the transcription units that bear C-rich transcripts of 41 bp tandem repeats (Solovei et al. 1995; Deryusheva et al. 2007). In the confocal sections of stained chicken and Japanese quail oocyte nuclei, the domains enriched with hnRNP K were observed (Fig. 5c, d). In chicken GVs, the bright foci containing hnRNP K epitopes recognized by 3С2 antibody were clearly revealed in the loops near the centromeric region on the long arm of chromosome 3 and in the subtelomeric regions of chromosomes 1 and 2 that corresponds well to the data on GV spreads. In some microchromosomes, RNP-matrixes of lateral loops enriched with hnRNP K also appeared as brightly stained domains. However, we have not detected any extrachromosomal domains enriched with hnRNP K. The fluorescence intensity level of most of the simple loops and the nucleoplasm after staining with Ab 3С2 was equivalent (Fig. 5c, d).
a In avian oocyte nuclei, splicing factor SC35 is concentrated in the RNP-matrix of complex giant loops of lampbrush chromosomes. Double staining of intact chicken (G. gallus domesticus) germinal vesicle with antibody against SR-protein SC35 (red) and nucleic acid specific fluorescent dye Sytox green (green). Arrows point to nuclear domains enriched with splicing factors. Related Supplementary materials—Video 5. b Distribution of TMG-capped small nuclear RNAs (red) in lateral loops of lampbrush chromosomes in intact Japanese quail (C. coturnix japonica) germinal vesicle. c–d Immunofluorescent staining of intact chicken oocyte nuclei with antibody 3С2 against protein K of heterogeneous nuclear RNPs (red) followed by Sytox green staining (green). Arrows point to nuclear domains enriched with hnRNP K. Maximum projection views of XYZ confocal stacks are shown. Scale bars a, c, d 50 μm, b 10 μm
Avian germinal vesicles versus amphibian germinal vesicles
The extrachromosomal domains in the oocyte nucleus were compared between avian and amphibian species. As expected from many previous findings (Gall et al. 2004; Sommerville 2010), in the Xenopus GVs we observed thousands of extrachromosomal nucleoli, approximately 100 of the smaller spherical bodies and several thousand B-snurposomes (the smallest bodies; Fig. 2d). A similar distribution of nuclear structures was typical for the oocytes of other frogs, namely P. ridibundus and P. lessonae (Fig. 2e).
In contrast, in the chicken, Japanese quail and chaffinch GVs isolated from large oocytes, we observed no extrachromosomal RNA-enriched structures determined by Sytox green staining (Fig. 2a, c; Video 1 and 2, Supplementary). Compared to those of amphibians, the large oocytes of sexually mature birds were devoid of any nucleoli, which corresponds to the earlier findings (Greenfield 1966; Gaginskaia and Chin 1980; Gaginskaia 1972; Gaginskaia and Gruzova 1975). The rest of the nuclear space was also free of any NA-enriched spheres or clusters of granules. The only exceptions were the rock pigeon GVs where several Sytox green-positive spherical bodies were detected in the series of confocal XYZ images (Fig. 2b). A detailed description of these remarkable extrachromosomal nuclear organelles of pigeon oocyte is presented elsewhere (Khodyuchenko et al. 2012). Confocal images also clearly demonstrate that in avian GVs, the previously described centromere-associated protein bodies (Gaginskaya and Gruzova 1969; Khutinaeva et al. 1989; Saifitdinova et al. 2003) do not contain RNA or DNA and appear as ‘black holes’ in nuclei stained for nucleic acids (Fig. 2f; Video 2, Supplementary). In larger avian oocyte nuclei, when the chromosomes condense into a smaller volume from 0.07 % down to 0.015 % of the nuclear space and RNP complexes are released from the lateral loops, no NA-containing structures other than chromosomes were detected in the chicken, quail or chaffinch GVs (Fig. 3c, d).
Distribution of splicing factors in avian germinal vesicles
To characterize the spatial organisation of the RNA splicing machinery in avian oocyte nuclei, we stained and imaged 3D-preserved GVs with antibodies against spliceosome components (Fig. 5a, b). The staining of the RNP-matrix in most of the transcriptionally active lateral loops and the nucleoplasm with antibodies against splicing factors (Fig. 5b) strongly corresponds with the immunofluorescent staining data of avian LBC spreads (Hutchison 1987; Krasikova et al. 2004). In chicken, chaffinch and pigeon oocyte nuclei, we observed a speckled pattern with antibodies against the SR-protein SC35 (Fig. 5a). In comparison with the GVs of amphibians, no extrachromosomal bodies containing splicing factor SC35 were detected in the GVs of the studied avian species during the stage of either the most intensive transcription or karyosphere formation.
Here we for the first time demonstrate that the highest concentration of SC35 splicing factor is typical for distinct chromosome-associated nuclear domains up to 15 μm in size that correspond to the so-called complex loops (terminal giant and interstitial lumpy loops; Fig. 5a, Video 5, Supplementary). These giant loops are usual for chicken, rock pigeon and chaffinch lampbrush chromosomes and are characterized by a complex morphology of the RNP-matrix (Khutinaeva et al. 1989; Chelysheva et al. 1990; Saifitdinova et al. 2003). The detailed description of molecular composition and organisation of complex loops of avian LBCs will be published elsewhere. In some GVs, patches of RNP-aggregates were also found around the lateral loops with the RNP-matrix enriched with SC35 protein.
Absence of Cajal bodies in nuclei of late-stage avian oocytes
We show here that in contrast to the Xenopus GVs, chicken, Japanese quail and chaffinch oocyte nuclei at the late stages of growth do not contain any extrachromosomal bodies enriched with 2,2,7-trimethylguanosine (TMG) cap of snRNAs or Sm proteins of snRNPs (Fig. S1). Moreover, we found that the GVs of these species do not have intranuclear organelles that accumulate coilin, which is a marker component of Cajal bodies (Morris 2008). Nucleolated cells of the follicular epithelium that coats avian oocytes comprise one or two nucleoli (Fig. 6c), several CBs (Fig. 6d) and multiple SC35-domains. These findings and positive controls signify that CBs do not form in the nuclei of large oocytes of these avian species. At the same time, the pigeon GV contains multiple free coilin-enriched spherical CB-like organelles that are characterized in detail by Khodyuchenko et al. (2012).
Germinal vesicles from 1-month-old Japanese quail (C. coturnix japonica) females comprise one or two nucleoli and several Cajal bodies. a Immunofluorescent staining of whole mount collagenase-treated oocyte with antibody against fibrillarin (green). Arrow points to nucleolus. b Transmitted light image of small oocyte showing one nucleolus (arrowhead). c Nucleolated cells of follicular epithelium (left) stained by antibody against NO38 (red) and corresponding transmitted light image (right). Arrows point to nucleoli. d–f Immunofluorescent staining of whole mount collagenase-treated quail oocytes with antibodies against coilin (d) and Sm-core proteins of small nuclear RNPs (e–f). Arrows point to coilin-positive body (d) and snRNP-containing bodies (e–f). Scale bars 10 μm
In contrast to the large growing oocytes of egg-laying females, germinal vesicles from non-adult females of chicken and Japanese quail contained one or two prominent nucleoli (Fig. 6a, b); the data correspond with the previous findings (Greenfield 1966; Chin et al. 1979). We found that in the small oocyte nuclei of 1-month-old chicken and Japanese quail females, there were several CB-like organelles (up to eight entities), which were detected with antibodies against coilin and small nuclear RNPs (Fig. 6d–f).
Absence of histone locus bodies and inactivation of histone gene cluster on chicken lampbrush chromosomes
Recently, coilin-enriched organelles in late Xenopus oocytes were classified as histone locus bodies that accumulate U7 snRNA (Nizami et al. 2010). We report here that large growing oocytes of the avian species studied so far lack both chromosome-associated and extrachromosomal HLBs. HLBs form in association with replication-dependent histone genes upon their transcriptional activation (Nizami et al. 2010). We aimed to examine the precise position and functional activity of the main histone gene cluster in growing avian oocytes using chicken oocytes as an example.
It is known that in the chicken genome 39 out of 44 histone genes form a cluster approximately 110 kbp long (Takami et al. 1996) (Fig. 7a). Using Basic Local Alignment Search Tool (BLAST) resources (Assembled RefSeq Genomes), we found that in the assembled sequences of the chicken genome (build 2.1) most of the histone genes are clustered in the 49.93–50.06 Mbp region of the chromosome 1 short arm (Fig. 7b). For high-resolution mapping of the histone gene locus, a BAC clone WAG115D22 (∼50 Mbp) containing genes for H2A and H2B was selected from the Wageningen chicken BAC library (Crooijmans et al. 2000). The presence of histone genes in this BAC clone was verified by a PCR with the primers H2BR and H2BF, which are specific to a conserved region of Н2В gene. A fragment of the expected length (230 bp) was amplified by PCR from both the chicken genomic DNA and DNA of WAG115D22.
Organisation of histone gene cluster in the chicken (G. gallus domesticus) genome and its physical mapping on lampbrush chromosomes. a Structure of histone gene cluster in the chicken genome (from Takami et al. 1996, with modifications). b 49.93–50.06 Mb region of GGA1 sequence assembly showing histone gene cluster (www.ncbi.nih.gov). c Physical mapping of BAC clone WAG115D22 (red fluorescence) containing histone genes on chicken lampbrush chromosome 1 (arrows) by DNA/DNA fluorescent in situ hybridization. d DNA/RNA fluorescent in situ hybridization of BAC clone WAG053E23 (LEI0071) (red) together with BAC clone WAG115D22 (green) on chicken lampbrush chromosome 1. Note the absence of signal from BAC WAG115D22 (arrows). Scale bars 30 μm. e Relative positions of fragments cloned in BAC WAG115D22, BAC WAG053E23 (LEI0071) and marker structures are shown on cytological chromomere map of chicken lampbrush chromosome 1
The high-resolution physical mapping of the histone gene cluster was performed on micro-surgically isolated chicken lampbrush chromosomes by FISH. A DNA fragment of the BAC clone WAG115D22 containing histone genes was localized to a region ‘C’ of the short arm of chicken lampbrush chromosome 1 (Fig. 7c, e). The cluster of histone genes occupies one DAPI-positive chromomere; there were no signals in the transcriptionally active lateral loops arising from this chromomere. When FISH with the same probe was performed according to the DNA/RNA hybridisation protocol, no signals from WAG115D22 were detected on chicken LBC 1 (Fig. 7d). Another probe that contained sequences transcribed during the lampbrush stage (ВАС WAG053E23) was added to hybridisation mix for a positive control. The added probe efficiently hybridised to nascent transcripts on a pair of small lateral loops on lampbrush chromosome 1 (Fig. 7d, e), which indicates that the RNA integrity was maintained in the preparations used. Thus in contrast to many other sequences, the histone gene cluster is not transcribed on chicken lampbrush chromosomes.
Although no spheres were observed in association with chicken LBC1 on spread preparations (Chelysheva et al. 1990), it could not be ruled out that during spreading of avian GV content spherical organelles detach from lampbrush chromosomes. In frogs and newts, previously described lampbrush chromosome-associated histone locus bodies can be detected in 3D preserved germinal vesicles by Sytox green staining (Fig. 2g). A morphological analysis of intact chicken GVs stained with Sytox green did not reveal any NA-enriched nuclear bodies associated with the chromomere containing histone gene cluster according to a mapped position of WAG115D22 (Video 6, Supplementary).
In summary, the general organisation of RNA processing machinery in avian germinal vesicles is not identical to that of amphibian GVs (Fig. 8). Coilin-positive Cajal bodies do not form inside the interchromatin space of GVs in all avian species studied with the only exception of pigeon GVs. Similarly, HLBs do not appear in the large oocytes of the studied species of birds. The oocyte nucleus of an egg-laying bird is also characterized by the absence of interchromatin granule clusters (Fig. 8). Instead, in the oocyte nuclei of chicken, rock pigeon and chaffinch, SC35-domains are represented by LBC-associated lumpy and terminal giant loops and their derivates (patches) in the nucleoplasm.
A scheme of organisation of growing oocyte nuclei (germinal vesicles, GVs) at the lampbrush stage in Xenopus and four species of birds (Japanese quail, chicken, chaffinch and pigeon) based on previous findings and novel data. In addition to lampbrush chromosomes, Xenopus GVs comprise amplified nucleoli (blue), coilin-positive extrachromosomal bodies and histone locus bodies (red), as well as interchromatin granule clusters (green) (reviewed in Gall et al. 2004; Nizami et al. 2010). In contrast, avian nucleoli-free GVs lack histone locus bodies and Cajal bodies with the only exception of pigeon GV. Pigeon oocyte nucleus contains multiple non-canonical Cajal bodies (red) (Khodyuchenko et al. 2012). In avian oocyte nuclei, SC35-domains are represented by chromosome-associated terminal giant and lumpy loops (green domains). Prominent protein bodies (grey) associated with centromere regions of chaffinch and pigeon lampbrush chromosomes (Gaginskaya and Gruzova 1969; Khutinaeva et al. 1989; Saifitdinova et al. 2003) are also shown
Discussion
Here, we present the results of our exploration into the 3D organisation of oocyte nuclei with special emphasise on RNA processing machinery in four species of birds. Important aspect of avian oocyte nuclear compartmentalisation is the absence of a number of evolutionarily conserved nuclear compartments. In contrast to somatic cells that have continuously active housekeeping genes, GVs have variable levels of activity of nucleolus organiser and histone genes and can be used as a favorable model to investigate the determinant variations in nuclear body formation.
We established for certain that chicken, Japanese quail and chaffinch germinal vesicles from large oocytes of egg-laying females are characterized by the absence of an essential subnuclear structure—the Cajal body (Fig. 8). Similarly, transcriptionally active nuclei of Xenopus late-stage oocytes lack canonical CBs (Nizami et al. 2010). The lack of CBs in the large oocytes from the three species of birds examined in this study is not due to the lack of nucleoli because the pigeon nucleolus-free oocyte has heterogeneous CB-like organelles (Fig. 8) (Khodyuchenko et al. 2012). If splicing events occur on the lateral loops of lampbrush chromosomes, how do splicing snRNPs mature in the absence of CBs? Despite the crucial role for CBs in the spliceosome cycle (Staněk and Neugebauer 2006; Morris 2008), proper modifications of spliceosome snRNAs can be completed even in the absence of CBs (Deryusheva and Gall 2009). Thus, in avian GVs lacking CBs, nuclear steps of snRNA maturation can occur freely in the nucleoplasm or in the small nucleoplasmic granules.
The CBs in the nucleolated oocytes of sexually immature birds are described here for the first time. It is possible that in chicken, quail and chaffinch ovaries, oocytes bearing coilin-positive nuclear bodies and nucleoli are eliminated. The more likely alternative is that both nucleoli (Chin et al. 1979) and CBs disintegrate in later staged oocytes.
Strikingly, avian GVs seem to be the only known example of transcriptionally active nuclei without functional HLBs, both chromosome associated and extrachromosomal (Fig. 8). Accordingly, the histone gene cluster located on chicken chromosome 1 is not transcribed during the lampbrush stage at least with respect to the H2A and H2B genes. Thus the inactivation of histone genes in chicken oocytes is accompanied by the non-appearance of any HLBs or associated CBs in this region. It can be suggested that, in addition to ribosomal RNA (Gaginskaya and Gruzova 1969), the cells of the follicular epithelium could supply the oocyte with required pool of histone gene mRNA, which would be stored until the early stages of embryogenesis. Accordingly, in addition to nucleoli (Gilbert et al. 1983), follicular ovary cells of different bird species contain at least one CB and one HLB. On the other hand, histone genes mRNAs could be synthesized in early oocytes and stored inside the growing oocyte until maturation.
It seems that in common with nucleoli and histone locus bodies, many other nuclear domains are associated with certain chromosome loci, but this association is still not well-established. According to the modern notions, nuclear domains that concentrate RNA-binding proteins could be structured by stable long non-coding RNAs (Shevtsov and Dundr 2011; Tripathi and Prasanth 2011). In amphibian and avian lampbrush chromosomes, a number of lateral loops accumulate specific sets of proteins in their RNP-matrixes and form at specific loci (Solovei et al. 1995; Abbott et al. 1999; Deryusheva et al. 2007; Morgan 2007). Such transcription units could correspond to nuclear domains highly enriched with specific sets of RNA-binding proteins and nascent transcripts; these domains in their turn could have somatic equivalents.
For instance, the hnRNP K protein associates with the C-rich transcripts of 41 bp tandem repeats on a number of lateral loops of chicken and Japanese quail lampbrush chromosomes (Deryusheva et al. 2007). In Galliform oocytes, the transcription units of 41 bp tandem repeats producing RNA that is capable of interacting with hnRNP K/J appear as chromosome-associated nuclear domains. Thus, the formation of domains enriched with hnRNP K in Galliform GVs is triggered by transcription of 41 bp tandem repeats. It can be summarised that the nuclear domains enriched with hnRNP K in Galliform GVs are similar to nuclear stress bodies that form at the transcription sites of satellite III in human cells. In fact, both types of domains concentrate transcripts of tandem/satellite DNA repeats in complex with specific set of RNP proteins and contain active form of RNA polymerase II (Jolly and Lakhotia 2006; Deryusheva et al. 2007).
Avian GVs lack structures resembling extrachromosomal interchromatin granule clusters (splicing speckles), which participate in dynamics of spliceosome components (Tripathi and Prasanth 2011). Instead, multiple SC35-domains were found here to correspond to the complex giant loops (Fig. 8), which were previously shown to form in terminal or interstitial regions of lampbrush chromosomes (Khutinaeva et al. 1989; Chelysheva et al. 1990; Saifitdinova et al. 2003; reviewed in Gaginskaya et al. 2009). In contrast to simple lateral loops, the terminal giant loops are devoid of elongating form of RNA polymerase II (Saifitdinova et al. 2003). The accumulation of SC35 splicing factor in the RNP-matrix of terminal giant and lumpy loops of avian LBCs if compared with the nucleoplasm is reported here for the first time.
Recently, we have demonstrated that one of these complex loops on chicken lampbrush chromosome 2 (so-called lumpy loop) forms at the locus of the non-coding tandem repeat LL2R (lumpy loop 2 repeat); RNP-matrix of the lumpy loops accumulated non-coding transcripts of LL2R repeat (Krasikova et al. 2010). Nascent transcripts of LL2R repeat associated with transcription unit on LBC2 are supposed to be involved into formation of SC35-domains in chicken oocyte nucleus in a way analogous to Nuclear Enriched Abundant Transcript 1—a long non-coding RNA accumulated in paraspeckles in mammalian species (Tripathi and Prasanth 2011). The complex loops with a high concentration of splicing factors and non-coding RNA on avian lampbrush chromosomes extend the list of nuclear domains that accumulate specific sets of RNP complexes at certain chromosomal loci. It is evident that formation of such depot in GVs is nucleated by active transcription of non-coding tandem repeats, which strongly supports a model of RNA-mediated nuclear domain formation (Shevtsov and Dundr 2011).
In summary, the nucleus of the growing avian oocyte has a unique organisation of RNA processing machinery. Exploration of the three-dimensional structure of giant nucleus of avian oocyte in combination with cytogenetic analysis of the loci of nuclear domain formation significantly reinforces the concept of eukaryotic nucleus compartmentalization.
Abbreviations
- 3D:
-
Three-dimensional
- Ab:
-
Antibody
- BAC:
-
Bacterial artificial chromosome
- CBs:
-
Cajal bodies
- CLSM:
-
Confocal laser scanning microscopy
- DAPI:
-
4′,6-diamidino-2-phenylindole
- FISH:
-
Fluorescent in situ hybridization
- GGA:
-
Chicken (Gallus gallus) chromosome
- GV:
-
Germinal vesicle
- HLBs:
-
Histone locus bodies
- hnRNPs:
-
Heterogeneous nuclear ribonucleoproteins
- LBCs:
-
Lampbrush chromosomes
- Mb:
-
Megabase
- PBS:
-
Phosphate-buffered saline
- PFA:
-
Paraformaldehyde
- SC35:
-
Splicing component (factor) 35 kDa
- snRNA:
-
Small nuclear RNA
- SR-protein:
-
Serine/arginine-rich protein
References
Abbott J, Marzluff W, Gall JG (1999) The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell 10:487–499
Bogolyubov D, Parfenov V (2008) Structure of the insect oocyte nucleus with special reference to interchromatin granule clusters and cajal bodies. Int Rev Cell Mol Biol 269:59–110
Callan HG (1986) Lampbrush chromosomes. Molecular Biology, Biochemistry and Biohysics 36:1–254
Callan HG, Lloyd L (1960) Lampbrush chromosomes of crested newts Triturus cristatus (Laurenti). Phil Trans R Soc Lond B 243:135–219
Chelysheva LA, Solovei IV, Rodionov AV, Yakovlev AF, Gaginskaya ER (1990) The lampbrush chromosomes of the chicken. The cytological map of macrobivalents Tsitologiia 32:303–316
Chen F, Gonzalez JM, Dustman WA, Moran MA, Hodson RE (1997) In situ reverse transcription, an approach to characterize genetic diversity and activities of prokaryotes. Appl Environ Microbiol 63(12):4907–4913
Chin SH, Gaginskaya ER, Kalinina EI (1979) Peculiarities of oogenesis in the chicken. I Extrafollicular period in the development of oocytes Ontogenes (Russ) 10:340–349
Committee for the Update of the Guide for the Care and Use of Laboratory Animals (2011) Guide for the Care and Use of Laboratory Animals, vol 8. The National Academic Press, Washington DC, p 220
Crooijmans RP, Vrebalov J, Dijkhof RJ, van der Poel JJ, Groenen MA (2000) Two-dimensional screening of the Wageningen chicken BAC library. Mamm Genome 11:360–363
De La Fuente R (2006) Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol 292(1):1–12
Deryusheva S, Gall JG (2009) Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 20(24):5250–5259
Deryusheva S, Krasikova A, Kulikova T, Gaginskaya E (2007) Tandem 41-bp repeats in chicken and Japanese quail genomes: FISH mapping and transcription analysis on lampbrush chromosomes. Chromosoma 116:519–530
Gaginskaia ER (1972) The nuclear structures in oocytes of adult birds. I Chromosome behavior during the oocyte cytoplasm growth Tsitologiia 14:426–432
Gaginskaia ER, Chin SH (1980) Peculiarities of oogenesis in the chicken. II Follicular period in oocyte development Ontogenes (Russ) 11:13–221
Gaginskaia ER, Gruzova MN (1975) Detection of the amplified rDNA in ovarial cells of some insects and birds by hybridization in situ. Tsitologiia 17:1132–1137
Gaginskaya E, Kulikova T, Krasikova A (2009) Avian lampbrush chromosomes: a powerful tool for exploration of genome expression. Cytogenet Genome Res 124:251–267
Gaginskaya ER, Gruzova MN (1969) Peculiarities of oogenesis in the chaffinch. Tsitologiia 11:1241–1251
Galkina S, Deryusheva S, Fillon V, Vignal A, Crooijmans R, Groenen M, Rodionov A, Gaginskaya E (2006) FISH on avian lampbrush chromosomes produces higher resolution gene mapping. Genetica 128(1–3):241–251
Gall JG, Wu Z (2010) Examining the contents of isolated Xenopus germinal vesicles. Methods 51:45–51
Gall JG, Wu Z, Murphy C, Gao H (2004) Structure in the amphibian germinal vesicle. Exp Cell Res 296:28–34
Gilbert AB, Perry MM, Waddington D, Hardie MA (1983) Role of atresia in establishing the follicular hierarchy in the ovary of the domestic hen (Gallus domesticus). J Reprod Fertil 69:221–227
Greenfield ML (1966) The oocyte of the domestic chicken shortly after hatching, studied by electron microscopy. J Embryol Exp Morphol 15:297–316
Hutchison N (1987) Lampbrush chromosomes of the chicken. Gallus domesticus J Cell Biol 105:1493–1500
Jolly C, Lakhotia SC (2006) Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res 34:5508–5514
Khodyuchenko T, Gaginskaya E, Krasikova A (2012) Non-canonical Cajal bodies form in the nucleus of late stage avian oocytes lacking functional nucleolus. Histochem Cell Biol 138:57–73
Khutinaeva MA, Kropotova EV, Gaginskaia ER (1989) The characteristics of the morphofunctional organization of the lampbrush chromosomes from the oocytes of the rock dove. Tsitologiia 31(10):1185–1192
Krasikova A, Deryusheva S, Galkina S, Kurganova A, Evteev A, Gaginskaya E (2006) On the positions of centromeres in chicken lampbrush chromosomes. Chromosome Res 14:777–789
Krasikova A, Kulikova T, Saifitdinova A, Derjusheva S, Gaginskaya E (2004) Centromeric protein bodies on avian lampbrush chromosomes contain a protein detectable with an antibody against DNA topoisomerase II. Chromosoma 113:316–323
Krasikova AV, Vasilevskaya EV, Gaginskaya ER (2010) Chicken lampbrush chromosomes: transcription of tandemly repetitive DNA sequences. Russ J Genet 46(10):1173–1177
Kropotova EV, Gaginskaya ER (1984) Lampbrush chromosomes from the Japanese quail oocytes. Tsitologiia 26:1008–1015
Kumaran RI, Thakar R, Spector DL (2008) Chromatin dynamics and gene positioning. Cell 132:929–934
Lemm I, Girard C, Kuhn AN et al (2006) Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 17:3221–3231
Lerner MR, Andrews NC, Miller G, Steitz JA (1981) Two small RNAs encoded by Epstein–Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A 78(2):805–809
Liu J-L, Buszczak M, Gall JG (2006) Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res 14:465–475
Macgregor HC, Sessions SK, Arntzen JW (1990) An integrative analysis of phylogenetic relationships among newts of the genus Triturus (family Salamandridae), using comparative biochemistry, cytogenetics and reproductive interactions. J Evol Biol 3:329–373
Maslova A, Krasikova A (2012) Nuclear actin depolymerization in transcriptionally active avian and amphibian oocytes leads to collapse of intranuclear structures. Nucleus 3:300–311
Maslova AV, Krasikova AV (2011) Spatial arrangement of macro-, midi-, and microchromosomes in transcriptionally active nuclei of growing oocytes in birds of order Galliformes. Cell and Tissue Biol 5:281–293
Matunis MJ, Michael WM, Dreyfuss G (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol 12(1):164–171
Morgan GT (2002) Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function. Chromosome Res 10:177–200
Morgan GT (2007) Localized co-transcriptional recruitment of the multifunctional RNA-binding protein CELF1 by lampbrush chromosome transcription units. Chromosome Res 15:985–1000
Morgan GT (2008) Working with oocyte nuclei: cytological preparations of active chromatin and nuclear bodies from amphibian germinal vesicles. Methods Mol Biol 463:55–66
Morris GE (2008) The Cajal body. Biochim Biophys Acta 1783:2108–2115
Nizami ZF, Deryusheva S, Gall JG (2010) Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harb Symp Quant Biol 75:313–320
Saifitdinova A, Derjusheva S, Krasikova A, Gaginskaya E (2003) Lampbrush chromosomes of the chaffinch (Fringilla coelebs L.). Chromosome Res 11:99–113
Shevtsov SP, Dundr M (2011) Nucleation of nuclear bodies by RNA. Nat Cell Biol 13:167–173
Solovei I, Macgregor H, Gaginskaya E (1995) Single stranded nucleic acid binding structures on chicken lampbrush chromosomes. J Cell Sci 108:1391–1396
Sommerville J (2010) Using oocyte nuclei for studies on chromatin structure and gene expression. Methods 51:157–164
Staněk D, Neugebauer KM (2006) The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 115:343–354
Takami Y, Higashio M, Fukuoka T, Takechi S, Nakayama T (1996) Organization of the chicken histone genes in a major gene cluster and generation of an almost complete set of the core histone protein sequences. DNA Res 30:95–99
Telenius H, Carter NP, Bebb CE, Nordenskjöld M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13(3):718–725
Tripathi V, Prasanth KV (2011) Cell Nucleus. eLS. doi:10.1002/9780470015902.a0001337.pub2
Zlotina A, Galkina S, Krasikova A, Crooijmans RPMA, Groenen MAM, Gaginskaya E, Deryusheva S (2012) Centromere positions in chicken and Japanese quail chromosomes: de novo centromere formation versus pericentric inversions. Chromosome Res. doi:10.1007/s10577-012-9319-7
Acknowledgements
The authors gratefully acknowledge Richard Crooijmans and Martin Groenen (the Wageningen University) for providing BAC clones from the Wageningen chicken BAC-library. The authors are grateful to Robert Hock (University of Wuerzburg) and Tatiana Kulikova (Saint-Petersburg State University) for very helpful methodological advices. We especially thank Herbert Macgregor and Garry Morgan for helpful suggestions for improvement of the manuscript. This investigation was supported by Russian Foundation for Basic Research (Grant #12-04-01807-а) and Federal Grant-in-Aid Program «Human Capital for Science and Education in Innovative Russia» (Agreement #2012-1.2.2-12-000-1013-026). The authors acknowledge Saint-Petersburg State University for a research grant #1.38.66.2011 and the Core Facility ‘CHROMAS’ (SPbSU) for an access to equipment.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Herbert Macgregor
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Absence of extrachromosomal bodies containing small nuclear RNPs in Japanese quail (Coturnix coturnix japonica) oocyte nucleus at the lampbrush stage. Immunofluorescent staining of intact quail oocyte nucleus with antibody Y12 against symmetrical dimethylarginine of Sm-proteins of small nuclear RNPs. Only lampbrush chromosomes are visible on sequential XYZ confocal 5 μm stacks of whole oocyte nucleus. Scale bar 20 μm. (JPEG 175 kb)
Video 1
Intact Japanese quail (Coturnix coturnix japonica) oocyte nucleus (germinal vesicle) after fluorescent staining with nucleic acid specific dye Sytox green was analysed with laser scanning confocal microscope. The overall 3D structure of the lampbrush chromosomes was not affected by nuclear fixation procedures. Movie showing sequential confocal sections through oocyte nucleus was constructed with LAS AF software (Leica-Microsystems CMS GmbH). (AVI 11445 kb)
Video 2
Intact chaffinch (Fringilla coelebs) oocyte nucleus (germinal vesicle) after fluorescent staining with nucleic acid specific dye Sytox green was analysed with laser scanning confocal microscope. Complex loops can be clearly distinguished in isolated germinal vesicle. Centromere protein bodies do not contain nucleic acids. Movie showing sequential confocal sections through oocyte nucleus was constructed with LAS AF software (Leica-Microsystems CMS GmbH). (AVI 12374 kb)
Video 3
3D reconstruction of chicken (G. gallus domesticus) oocyte nucleus (germinal vesicle), where 39 isosurfaces corresponding to individual lampbrush bivalents were computer-painted into different colours. Volume reconstruction and movie were made in Imaris software (Bitplane, AG). The image segmentation used a threshold that allows for the visualization of individual lampbrush chromosomes and partitioning of the nuclear volume into a number of isosurface objects that correspond to distinct lampbrush bivalents. Related image—Fig. 3b. (AVI 8841 kb)
Video 4
Estimated gravity centres positions of macro-, midi- and microchromosomes at the lampbrush stage (pink, blue and green dots correspondingly) marked on the 3D-reconstrution of Japanese quail (C. coturnix japonica) oocyte nucleus. Gravity centres were estimated by computer analysis of chromosome isosurfaces and superimposed on the volume reconstruction of nucleus stained with Sytox green (red fluorescence). Reconstructions and movie were made in Imaris software (Bitplane, AG). Related image—Fig. 3b. (AVI 2171 kb)
Video 5
3D reconstruction of isolated chicken (G. gallus domesticus) oocyte nucleus immunostained with antibody against splicing factor SC35. Isosurfaces were constructed around lampbrush bivalents stained with Sytox green (green) and intranuclear domains accumulating splicing factor SC35 (red) with the highest intensity of fluorescence. Reconstructions and movie were made in Imaris software (Bitplane, AG). Related image—Fig. 5a. (AVI 8399 kb)
Video 6
Fragment of isolated chicken (G. gallus domesticus) oocyte nucleus showing 3D-preserved lampbrush bivalent 1 (GGA1). Movie was constructed with LAS AF software (Leica-Microsystems CMS GmbH). (AVI 5732 kb)
Rights and permissions
About this article
Cite this article
Krasikova, A., Khodyuchenko, T., Maslova, A. et al. Three-dimensional organisation of RNA-processing machinery in avian growing oocyte nucleus. Chromosome Res 20, 979–994 (2012). https://doi.org/10.1007/s10577-012-9327-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-012-9327-7