Abstract
Background
There is increasing interest in the molecular profiling of tumour tissues in order to investigate alternative breast cancer (BC) therapies. However, the impact of genomic screening for druggable mutations with targeted gene panel sequencing (TGPS) in routine practice remains controversial.
Methods
This is a retrospective analysis of data from a genomic screening programme at our institution, in which we performed simplified TGPS for mutations in PIK3CA, AKT1, KRAS, NRAS, and BRAF in order to select patients for targeted therapy clinical trials. The genomes of archived samples of primary (PT) and/or metastatic (MT) tumours from advanced BC patients were analysed with MassARRAY technology (Sequenom MassARRAY, OncoCarta v1.0). The level of PTEN expression was assessed by immunohistochemistry. The primary endpoint was to identify the proportion of BC patients with PI3 K and MAPK alterations who were included in clinical trials using targeted therapies against these pathways.
Results
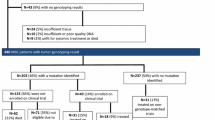
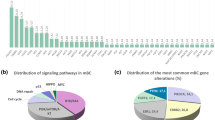
Two hundred and fifteen metastatic BC patients (65 PT and 168 MT) were included. Fifty-two patients (24.19 %) were enrolled in tailored clinical trials, of whom 29 (55.77, 13.49 % of all patients screened) harboured mutations targeted by the study drug. Moreover, 12 wild-type patients out of the 215 (5.58 %) were included in the clinical trials for which mutation analysis was an inclusion criteria. All the patients received drugs targeting the PI3K-AKT pathway and only two were given combinations directed against the PI3K and MAPK pathways. PI3KCA mutations were present in 33.7 % (61/181) of the patients, 45.83 % in PTs and 29.32 % in MTs. AKT1 mutations were detected in 5.48 % (8/146) of patients and PTEN loss in 34.67 % (52/150). KRAS, NRAS, and BRAF mutations were present in 12.06, 5.67, and 3.18 % of patients, respectively.
Conclusions
Genomic screening with a simplified TGPS is feasible, and was used to identify 13.49 % of patients who were included in clinical trials using targeted therapy against the mutations they harboured; PI3KCA mutations were the most frequent aberration in our series.


Similar content being viewed by others
Abbreviations
- BC:
-
Breast cancer,
- TGPS:
-
Targeted gene panel sequencing,
- PT:
-
Primary tumour,
- MT:
-
Metastatic tumour,
- IHC:
-
Immunohistochemistry,
- ER:
-
Oestrogen receptor
- HER2:
-
Human epidermal growth factor receptor 2
- NGS:
-
Next generation sequencing
- PI3K:
-
Phosphoinositide 3-kinase
- MAPK:
-
Mitogen-activated protein kinase
- TNBC:
-
Triple-negative breast cancer
- LBC:
-
Luminal breast cancer
- FFPE:
-
Formalin-fixed paraffin-embedded
- FISH:
-
Fluorescence in situ hybridisation
- EGFR:
-
Epidermal growth factor receptor
- HE:
-
Haematoxylin and eosin
- PR:
-
Progesterone receptor
- SAP:
-
Shrimp alkaline phosphatase
- CGAN:
-
Cancer genome ATLAS network
- FGFR:
-
Fibroblast growth factor receptor
- AKT1:
-
V-akt murine thymoma viral oncogene homolog 1
- KRAS:
-
Kirsten rat sarcoma viral oncogene homolog
- NRAS:
-
Neuroblastoma ras viral (V-ras) oncogene homolog
- BRAF:
-
B-raf proto-oncogene, serine/threonine kinase
- PTEN:
-
Phosphatase and tensin homolog
- PIK3CA:
-
Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha
References
Arnedos M, Vicier C, Lefebvre C et al (2015) Precision medicine for metastatic breast cancer: limitations and solutions. Nature Rev 12:693–704
McCubrey JA, Steelman LS, Abrams SL et al (2006) Roles of the RAF/MEK/ERK and PI3 K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul 46:249–279
Cizkova M, Cizeron-Clairac G, Vacher S et al (2010) Gene expression profiling reveals new aspects of PIK3CA mutation in ERalpha-positive breast cancer: major implication of the Wnt signaling pathway. PLoS One 5(12):e15647
Baselga J (2011) Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist 16(Suppl 1):12–19
Lievre A, Bachet J, Le Corre D et al (2006) KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 66:3992–3995
Giltnane J, Balko J (2014) Rationale for targeting the RAS/MAPK pathway in triple negative breast cancer. Discov Med 17(95):275–283
Nguyen DX, Bos PD, Massagué J (2009) Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9(4):274–284
Jamal-Hanjani M, Quezada SA, Larkin J et al (2015) Translational implications of tumor heterogeneity. Clin Cancer Res 21(6):1258–1266
Gerlinger M, Rowan AJ, Horswell S et al (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Eng J Med 366(10):883–892
Ibarrola-Villava M, Fleitas T, Llorca-Cardeñosa MJ et al (2016) Determination of somatic oncogenic mutations linked to target-based therapies using MassARRAY technology. Oncotarget 7(16):22543–22555
Quail MA, Smith M, Coupland P et al (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina Miseq sequencers. BMC Genom 13:341
Andre F, Bachelot T, Commo F et al (2014) Comparative genomic hybridization array and DNA sequencing to direct treatment in metastatic breast cancer: a multicenter prospective trial (SAFIR01/UNICANCER). Lancet Oncol 15:267–274
Hollebecque A, Massard C, de Baere T et al (2013) Molecular screening for cancer treatment optimization (MOSCATO 01): a prospective molecular triage trial-interim results. J Clin Oncol 31:15
Commo F, Ferté C, Soria JC et al (2015) Impact of centralization on a CGH-based genomic profiles for precision medicine in oncology. Ann Oncol 26(3):582–588
Gonzalez-Angulo AM, Ferrer-Lozano J, Stemke-Hale K et al (2011) PI3 K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Ther 10(6):1093–1101
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A et al (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68:6084–6091
Lopez-Knowles E, O’Toole SA, McNeil CM et al (2010) PI3K pathway activation in breast cancer is associated with the basal like phenotype and cancer-specific mortality. Int J Cancer 126:1121–1131
CGAN (Cancer Genome ATLAS Network) (2012) Comprehensive molecular portraits of human breast tumors. Nature 490(7418):61–70
Pereira CB, Leal MI, De Souza CR et al (2013) Prognostic and predictive significance of MYC and KRAS alterations in breast cancer from women treated with neoadjuvant therapy. PLoS One 8(3):e60576
Koffa M, Malamoumitsi V, Agnantis N et al (1994) Mutational activation of k-ras oncogene in human breast-tumors. Int J Oncol 4(3):573–576
Tilch E, Seidens T, Cocciardi S et al (2014) Mutations in EGFR, BRAF and RAS are rare triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res Treat 143(2):385–392
de Dueñas EM, Hernández AL, Zotano AG et al (2014) Prospective evaluation of the conversion rate in the receptor status between primary breast cancer and metastasis: results from the GEICAM 2009-03 ConvertHER study. Breast Cancer Res Treat 143(3):507–515
Meric-Bernstam F, Frampton G, Ferrer-Lozano J et al (2014) Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther 13(5):1382–1389
Vasan N, Yelensky R, Wang K et al (2014) A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations ni primary and metastatic breast cancers: implications for clinical practice. Oncologist 19(5):453–458
Acknowledgments
This study was supported by grants from the Generalitat Valenciana (Prometeo 2015/005), the Ministerio de Salud Carlos III (PI12/02,767 and PI13/00606), and European FEDER Funds. MTM was funded by the Ministerio de Salud Carlos III on a Rio Hortega contract (CM12/00264). GR and MI-V were funded by the Ministerio de Salud Carlos III on a Miquel Servet II (CPII14-00013) and Sara Borell contract (CD15/00153), respectively. We would like to thank the expert personnel at the Genotyping and Epigenetics Laboratory at the Central Biomedical Research Unit (Unidad Central de Investigación de Medicina; UCIM) at the University of Valencia.
Author contributions
Study conception and design: ALL, AC, JAPF, and JMC. Data acquisition: OB, GR, and MI-V. Data analysis and interpretation: all authors. All the authors were also involved in drafting or critically revising the article for important intellectual content, and they approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors state that they have no potential conflict of interest.
Ethics approval
This research was performed in accordance with the Declaration of Helsinki and was approved by the ethics committee at the Hospital Clinico Universitario, Valencia.
Rights and permissions
About this article
Cite this article
Cejalvo, J.M., Pérez-Fidalgo, J.A., Ribas, G. et al. Clinical implications of routine genomic mutation sequencing in PIK3CA/AKT1 and KRAS/NRAS/BRAF in metastatic breast cancer. Breast Cancer Res Treat 160, 69–77 (2016). https://doi.org/10.1007/s10549-016-3980-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3980-z