Abstract
The biological subtype of breast cancer influences the selection of systemic therapy. Distinction between luminal A and B cancers depends on consistent assessment of Ki-67, but substantial intra-observer and inter-observer variability exists when immunohistochemistry (IHC) is used. We compared RT-qPCR with IHC in the assessment of Ki-67 and other standard factors used in breast cancer subtyping. RNA was extracted from archival breast tumour tissue of 769 women randomly assigned to the FinHer trial. Cancer ESR1, PGR, ERBB2 and MKI67 mRNA content was quantitated with an RT-qPCR assay. Local pathologists assessed ER, PgR and Ki-67 expression using IHC. HER2 amplification was identified with chromogenic in situ hybridization (CISH) centrally. The results were correlated with distant disease-free survival (DDFS) and overall survival (OS). qPCR-based and IHC-based assessments of ER and PgR showed good concordance. Both low tumour MKI67 mRNA (RT-qPCR) and Ki-67 protein (IHC) levels were prognostic for favourable DDFS [hazard ratio (HR) 0.42, 95 % CI 0.25–0.71, P = 0.001; and HR 0.56, 0.37–0.84, P = 0.005, respectively] and OS. In multivariable analyses, cancer MKI67 mRNA content had independent influence on DDFS (adjusted HR 0.51, 95 % CI 0.29–0.89, P = 0.019) while Ki-67 protein expression had not any influence (P = 0.266) whereas both assessments influenced independently OS. Luminal B patients treated with docetaxel-FEC had more favourable DDFS and OS than those treated with vinorelbine-FEC when the subtype was defined by RT-qPCR (for DDFS, HR 0.52, 95 % CI 0.29–0.94, P = 0.031), but not when defined using IHC. Breast cancer subtypes approximated with RT-qPCR and IHC show good concordance, but cancer MKI67 mRNA content correlated slightly better with DDFS than Ki-67 expression. The findings based on MKI67 mRNA content suggest that patients with luminal B cancer benefit more from docetaxel-FEC than from vinorelbine-FEC.
Similar content being viewed by others
Introduction
Biological subtyping of breast cancer is an integral part of the standard evaluation of patients diagnosed with breast cancer. Subtyping can be done with gene expression arrays [1], but the molecular subtypes are frequently approximated with immunohistochemistry (IHC) due to its wide availability and low cost. However, assays for cancer oestrogen receptor (ER), progesterone receptor (PgR) and human epidermal growth factor receptor-2 (HER2) expression by IHC have an up to 20 % risk for discordant or erroneous results [2, 3], and making a distinction between luminal A and luminal B breast cancer requires assessment with the proliferation marker Ki-67, which is prone to high intra- and inter-observer assessment variability [4, 5].
In this study, we compared assessment of breast cancer key biomarkers, ER, PgR, HER2 and Ki-67 quantitatively using RT-qPCR with their assessment using IHC or in situ hybridization as a part of the clinical routine in breast cancer subtyping and prediction of patient outcome. We hypothesized that quantifying Ki-67 with RT-qPCR might result in more robust outcome predictions. To our knowledge, few such comparative data are available.
Methods
Patients
The clinical data and breast tumour tissue samples were collected within the FinHer trial (identifier ISRCTN76560285), where 1010 women with axillary node–positive or high-risk axillary node-negative breast cancer were randomly assigned between October 2000 and September 2003 to receive either three cycles of docetaxel followed by three cycles of fluorouracil, epirubicin and cyclophosphamide (FEC) or three cycles of vinorelbine followed by three cycles of FEC [6, 7]. Breast tumour erbB2 (HER2) copy numbers were determined centrally by chromogenic in situ hybridization (CISH), and women with HER2-positive cancer (n = 232) had a second randomisation between nine weekly infusions of trastuzumab, given concomitantly with either docetaxel or vinorelbine, and similar chemotherapy without trastuzumab. After a median follow–up time of 62 months since randomisation, women assigned to docetaxel had better distant disease-free survival (DDFS, the primary objective) than those assigned to vinorelbine (HR 0.66, 95 % CI 0.49–0.91; P = 0.010) [6]. The absolute benefit in 5-year DDFS in favour of the docetaxel plus FEC regimen was 5.2 % (86.8 vs 81.6 %), and 3.3 % (92.6 vs 89.3 %) for overall survival (OS) across all biological subtypes [6].
Immunohistochemistry
Immunostaining for ER, PgR, HER2 and Ki-67 was performed on tissue sections cut from formalin-fixed, paraffin-embedded (FFPE) tumour tissue at the local pathology laboratories of the 17 study sites (all located in Finland) according to each laboratory’s standard procedures.
ER and PgR were considered positive when 10 % or more of the cancer cells stained positively. Ki-67 assays were analysed by estimating the proportion of positively staining cancer cell nuclei out of all cancer cell nuclei in the tissue section, and the result was provided as a percentage ranging from 0 to 100 %. For the present study, Ki-67 expression was considered positive when ≥20 % of cancer cell nuclei stained positively. Local pathologists interpreted the ER, PgR and Ki-67 immunostaining results, as per each institute’s standard practice.
Chromogenic in situ hybridization (CISH)
Tumours with a score of 2+ or 3+ (on a scale of 0 to 3+) for HER2 expression in IHC were further analysed for HER2 gene amplification by CISH in one of two central laboratories. The HER2 status was considered positive when six or more gene copies per nucleus were present. As in the original trial [6, 7], in the present study, cancer HER2 status was considered positive whenever CISH for HER2 was positive, and negative whenever CISH was negative, regardless of the degree of HER2 protein expression in IHC.
RT-qPCR
After pathologic confirmation of representativeness of the tissue sections for presence of cancer, a single whole-face 10-μm-thick slice from each FFPE tumour block was processed with the RNXtract® RNA extraction kit (BioNTech Diagnostics GmbH, Mainz) using a magnetic particle-based assay (Supplemental file 1A). RT-qPCR was done with the MammaTyper® kit (BioNTech Diagnostics GmbH, Mainz) for ESR1, PGR, ERBB2 and MKI67, and the two reference genes B2 M and CALM2 on a Versant kPCR system (Siemens, Erlangen, Germany) by applying one cycle of primer-specific reverse transcription followed by 40 cycles of nucleic acid amplification (Supplemental file 1B). The median quantification cycle (Cq) for each of the four genes of interest (GOI) were normalized against the two reference genes (REF) and presented as ΔΔCq values relative to the positive control, obtained after subtracting the ΔCq value of the positive control (pc) from the ΔCq of the sample (s) by the formula
To exclude a major influence of a varying tumour cell content for the assay results, sensitivity studies were undertaken similarly as previously reported [8]. A series of extreme cases with low content of invasive carcinoma and varying amount of DCIS were analysed before and after macrodissection and it could be confirmed that the TCC did not influence the final test result [9, Laible et al. submitted]. Therefore, a major influence of TCC on MKI67 mRNA expression can be excluded. Cut-offs for the markers ERBB2, ESR1 and PGR were defined in an independent technical cohort based on reference pathology IHC results. Prognostic and predictive value of MKI67 cut-offs had previously been analysed by testing objective cut-offs in 562 Affymetrix U133 A datasets from breast cancer patient cohorts having received either no systemic therapy, only endocrine treatment or chemo-endocrine regimen [10]. In view of these analyses, the MKI67 cut-off was set at the 3rd quartile of the normally distributed MKI67 expression data from 90 FFPE breast cancer reference tumour samples and thus ought to reflect a correlate to the standard Ki-67 cut-off at 20 % positively stained nuclei.
Definition of breast cancer biological subtypes
After defining each of the four biomarkers either positive or negative, the molecular subtype of each tumour was determined using a slightly modified version of the currently proposed IHC-based breast cancer molecular subtyping algorithm [1] (Supplemental File 1C). In brief, luminal A cancers were defined as having high ESR1 and/or PGR mRNA content and low ERBB2 and MKI67 content. Luminal B cancers were defined as having high cancer ESR1 and MKI67 content, or high ESR1 content but low PGR and ERBB2 content. Cancers with a high ERBB2 mRNA content were considered as HER2-positive cancers and were not further categorized into luminal and non-luminal (“enriched”) lesions. Triple-negative cancers consisted of cancers that had low ESR1, PGR and ERBB2 mRNA content irrespective of cancer MKI67 mRNA content.
The same scheme was used to categorize the cancers according to the IHC and CISH results, but using protein expression (at IHC) and the number of HER2 gene copies (at CISH) in place of cancer mRNA content. For example, cancers that were positive for ER and PgR (with ≥10 % of the nuclei that were positive in each staining), HER2 negative (by CISH) and had low Ki-67 (<20 % of nuclei stained positively at IHC) were considered luminal A cancers.
Statistical methods
The results were analysed according to a statistical analysis plan written and approved prior to the initiation of the study, and the RT-qPCR results were interpreted blinded to the clinical information. Kappa (κ) statistic numeric values are categorized into poor (≤0.2), fair (>0.2–0.4), moderate (>0.4–0.6), good (>0.6–0.8) and very good (>0.8) associations, and were used as a measure of positive percent agreement (PPA), negative percent agreement (NPA) and overall percent agreement (OPA). The tests are accompanied by their respective 95 % confidence intervals (95 % CI). A two-sided P value <0.05 was considered significant.
The primary clinical endpoint was DDFS, defined as the time period between the date of randomisation and the date of first distant metastasis or the date of death when death preceded detection of distant recurrence. Overall survival (OS) was defined as the time period between the date of randomisation and the date of death. Survival was analysed using the Kaplan–Meier method.
Univariable and multivariable Cox proportional hazards models were constructed to compare prognosis between groups and to study the interactions between variables. Hazard ratios (HRs) were calculated using a univariable Cox model. In multivariable Cox models, a backward selection procedure was used to adjust for the covariables.
Results
Patients
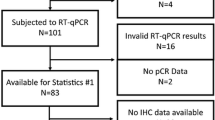
An RT-qPCR assay of ESR1, PGR, ERBB2 and MKI67 was successfully performed from breast cancer tissues of 769 (76 %) out of the 1010 patients entered to the FinHer trial. In the remaining 241 cases, cancer tissues were not available, and the tissue block did not consist mostly of cancer cells, or RNA extraction did not yield good-quality mRNA. We included in this study all 719 (71 % out of 1010) cases with successful RT-qPCR assay of the four genes and with IHC data available for subtyping. The inaccessibility rate to the tissue samples was similar across the study treatment arms (a modified CONSORT diagram shown in Supplemental file 1D). The characteristics of the patients and tumours included in the present study (Table 1) were similar to those of the entire FinHer trial cohort [7].
The median age of the patients at study entry was 50.9 years (range, 25.5–65.8). Tumours had a mean diameter of 26 mm ± 16 mm (6–150 mm), and the majority (n = 637, 88.6 %) had given rise to regional lymph node metastases at the time of the diagnosis. There were 511 (71.1 %) ER-positive, 395 (54.9 %) PgR-positive and 163 (22.7 %) HER2-positive cancers. After random allocation, 357 (49.7 %) patients were treated with docetaxel plus FEC, 362 (50.4 %) with vinorelbine plus FEC and 83 (50.9 %) of the 163 patients with HER2-positive cancer received trastuzumab. The median follow-up time after randomisation was 62 months, during which time period 112 patients had distant cancer recurrence and 62 died.
Concordance between mRNA and IHC assays
Tumour ESR1, PGR and ERBB2 mRNA content assessed by RT-qPCR and the corresponding protein expressions determined by IHC for ER and PgR and DNA amplification status by CISH for HER2 showed good concordance, whereas cancer MKI67 mRNA content and protein expression correlated only moderately well (Table 2). Many of the discordant cases between IHC and the mRNA assay had a high cancer MKI67 mRNA content, but despite this, <20 % of cancer cell nuclei stained positively with IHC (Fig. 1 ).
A scatterplot depicting the relation between tumour MKI67 mRNA content measured with RT-qPCR, and Ki-67 expression determined by immunohistochemistry (IHC). Vertical axis, tumour Ki-67 expression (IHC, %); horizontal axis, tumour relative MKI67 mRNA expression. The cut-off for positivity was 20 % in the Ki-67 protein assays (the horizontal line) and 34.8 in the MKI67 mRNA (qPCR) assays (the vertical line). Sections A and D depict the discordant cases, and sections B and C depict the concordant cases
Prognostic value of cancer MKI67 mRNA content and Ki-67 expression
Patients with low breast tumour MKI67 mRNA content or low (<20 %) Ki-67 expression had more favourable DDFS and OS as compared to those with high MKI67 mRNA content or Ki-67 expression. Each method produced roughly similar hazard ratios for DDFS and OS (Fig. 2).
In a multivariate Cox regression analysis where the type of chemotherapy (vinorelbine-FEC or docetaxel-FEC), the axillary nodal status (pN0, pN1, pN2 or pN3), tumour size (as a continuous variable), histological grade (as a continuous variable) and cancer MKI67 mRNA content (as a continuous variable) were entered as covariables, low tumour MKI67 mRNA content was independently associated with favourable DDFS (adjusted HR 0.51; 95 % CI, 0.29–0.90; P = 0.019) together with a negative axillary nodal status (P < 0.0001) and small cancer size (P = 0.006). A low cancer MKI67 mRNA content was also independently associated with favourable OS (adjusted HR 0.44; 95 % CI, 0.23-0.87; P = 0.018) in addition to the axillary nodal status (P = 0.003) and tumour size (P = 0.006). When Ki-67 protein expression was entered into the same models in place of cancer mRNA content, Ki-67 was not significantly associated with DDFS (P = 0.266), but when OS was selected as the endpoint, low cancer Ki-67 expression was associated with favourable survival (adjusted HR 0.43; 95 % CI, 0.24–0.77; P = 0.005) together with the axillary nodal status (P = 0.002) and small tumour size (P = 0.006).
Concordance of molecular subtyping with IHC and RT-qPCR
The method of Ki-67 assessment had substantial impact on making the distinction between luminal A and B cancers. Of the 189 cancers that were classified as luminal A by IHC/CISH, only 102 (54.0 %) were similarly classified, when MKI67 mRNA expression was used in place of Ki-67 protein staining with the 87 discordant cases being classified as either luminal B (n = 75, 39.7 %) or HER2 positive (n = 12, 6.4 %, Table 3). Of the 251 cancers that were classified as luminal B by IHC/CISH, 180 (71.7 %) were similarly classified using MKI67 mRNA expression, 48 (19.1 %) were classified as luminal A, 17 (6.8 %) as HER2 positive and 6 (2.4 %) as triple negative. Of the 156 and 294 tumours classified as luminal A and luminal B by RT-qPCR, respectively, 102 (65.4 %) and 180 (61.2 %) were classified as luminal A or B also with IHC/CISH.
Influence of cancer MKI67 mRNA expression-based and Ki-67 protein expression-based subtypes on outcome
There was no significant difference in DDFS or OS between patients treated with adjuvant docetaxel plus FEC and those treated with vinorelbine and FEC in the subsets with luminal A, HER2-positive or triple-negative breast cancer when each subtype was defined either with IHC/CISH or with RT-qPCR (DDFS and OS statistics for each subtype according to chemotherapy agent shown in Supplemental file 1E). Interestingly, when luminal B subtype was defined by MKI67 mRNA expression, patients treated with docetaxel plus FEC had significantly more favourable DDFS and OS as compared with those treated with vinorelbine plus FEC (for DDFS, HR 0.52, 95 % CI 0.29–0.94, P = 0.031; OS, HR 0.24, 95 % CI 0.09–0.65, P = 0.005). In contrast no significant difference in DDFS or OS was found, when the luminal B subtype was defined with Ki-67 protein expression (P > 0.10 for both analyses; Fig. 3).
Distant metastasis-free survival (panels a and b) and overall survival (panels c and d) of patients treated with adjuvant docetaxel plus FEC and those treated with vinorelbine plus FEC in the subset of patients with luminal B breast cancer. Panels a and c, the luminal B subtype was defined with MKI67 mRNA expression; panels b and d, the luminal B subtype was defined with Ki-67 protein expression
The type of adjuvant chemotherapy (tested docetaxel plus FEC vs vinorelbine plus FEC) had an independent influence on DDFS in the subset of patients who had luminal B cancer defined by cancer MKI67 mRNA content in a multivariable analysis (HR 0.44; 95 % CI 0.23–0.84, P = 0.013), together with cancer histological grade (tested as a continuous variable; HR 1.67, 95 % CI 1.03–2.72, P = 0.039) and tumour size (tested as a continuous factor; HR 1.02, 95 % CI 1.00–1.04, P = 0.026). Similarly, when OS was used as the end point in place of DDFS and the luminal B subtype was defined by cancer MKI67 mRNA content, docetaxel-containing chemotherapy was independently associated with favourable survival (HR 0.22; 95 % CI 0.08–0.60, P = 0.003) together with histological grade (HR 2.29, 95 % CI 1.15–4.57; P = 0.019), while tumour size lost its significance. Unlike MKI67 mRNA content, Ki-67 protein expression did not have independent influence on DDFS or OS in these models. When the luminal B subtype was defined with tumour MKI67 mRNA content, the interaction with the type of adjuvant chemotherapy given was significant (P = 0.040) when OS was selected as the end point, but not when DDFS was considered (P = 0.352). No interaction with either OS or DDFS and the type of adjuvant chemotherapy was present when the luminal subtype was defined with Ki-67 protein expression (P = 0.658 and 0.699, respectively).
Discussion
We approximated commonly used breast cancer biological subtypes using RT-qPCR and compared the results with the subtypes defined by IHC (and with CISH to detect HER2 amplification) within the framework of a large randomized clinical trial. The subtypes defined with each method agreed moderately well with most discrepancy occurring in the luminal B subtype. Both high cancer Ki-67 protein expression and high MKI67 mRNA content were associated with unfavourable DDFS and OS in a univariable analysis with approximately similar hazard ratios, but only tumour MKI67 mRNA content remained significant in a multivariable model for DDFS when both parameters were entered into the same model after a stepwise selection process of the covariables such as tumour size, nodal status, histological grade and the type of treatment given.
A key difference between luminal A and luminal B subtypes is a higher cell proliferation rate in the latter, which is often assessed by estimating the proportion of cancer cells that stain positively for Ki-67 after immunohistochemical staining. Interestingly, when the luminal B type was defined using cancer MKI67 mRNA content in place of Ki-67 expression assessed with immunohistochemistry, patients with luminal B breast cancer were found to benefit more from adjuvant docetaxel plus FEC than from adjuvant vinorelbine plus FEC, which association could not be detected when the luminal B breast cancer subtype was defined by Ki-67 protein expression with immunohistochemical staining.
Biological subtyping of breast cancer is the basis for selection of systemic cancer treatment [1]. Of the four biomarkers commonly used for this purpose, i.e. ER, PgR, HER2 and Ki-67, the assays for Ki-67 have turned out the most challenging ones to standardize and to make reproducible. For example, in a study carried out in a few leading pathology laboratories, there was substantial variability between the laboratories in scoring of Ki-67 expression from shared breast cancer tissue slides stained with IHC, and attempts to reduce the interlaboratory variability were only partially successful [4]. In the present study, IHC staining for Ki-67 was done locally in many pathology laboratories using the institutional staining protocols and was assessed by many pathologists, whereas cancer MKI67 mRNA content was determined centrally in one laboratory. To reduce the potential variability in Ki-67 staining and scoring, we considered carrying out staining for Ki-67 also centrally, but due to the difficulties to standardize Ki-67 immunostaining even in leading laboratories and to establish a reference procedure [4], we preferred to use the Ki-67 staining results reported originally by the local laboratories from whole tumour tissue sections as the comparator for the MKI67 mRNA assay. Image analysis of Ki-67 from IHC stained slides is a promising method to improve the reproducibility of Ki-67 scoring from immunostained slides, but, to our knowledge, no standard parameter values for scoring of the nuclei as either positive or negative are available. To estimate how well the locally assessed Ki-67 assays done from whole tumour tissue sections might correlate with a centrally done Ki-67 assay, we analysed cancer Ki-67 expression from TMAs (as whole tumour sections were not available) containing tissue from 745 breast cancers using image analysis [11]. The median cancer Ki-67 expression turned out to be similar with image analysis and locally done IHC (19.7 and 20.0 %, respectively), and the two assays showed strong correlation (P < 0.0001, Spearman’s rho 0.633). These observations suggest that centrally done image analysis of Ki-67 might have resulted in similar conclusions had it been selected as the comparator assay in place of the local Ki-67 IHC assays.
The subtypes defined with MKI67 mRNA were associated with survival outcomes that agree well with the results obtained with IHC from other clinical trials [9, 10, 12–14]. Patients with the luminal A subtype had the best 5-year DDFS, patients with HER2 positive and triple-negative cancer had the least favourable outcomes, while patients with luminal B cancer had an outcome intermediate of these subtypes (see Supplement File 1E). These results are well in agreement despite slight dissimilarities in the definition of luminal B and HER2-positive subtypes between the trials.
Taxane-containing adjuvant regimens are effective in the treatment of early breast cancer but are associated with side effects, and therefore, methods to optimize patient selection for regimens that contain a taxane are needed. The current finding that patients with luminal B cancer have longer DDFS and OS when treated with docetaxel plus FEC as compared with vinorelbine plus FEC is supported by observations made by Jacquemier et al. and Nitz et al. who found that chemotherapy containing docetaxel was associated with a significant reduction in the risk of relapse in the subset of patients with luminal B breast cancer in the PACS 01 trial [13] and WSG-AGO EC-Doc trial [12], respectively. Both of these trials compared docetaxel-containing regimens with standard anthracycline-containing treatments. In the BCIRG 001 trial that compared docetaxel, doxorubicin and cyclophosphamide (TAC) versus fluorouracil, doxorubicin and cyclophosphamide (FAC) in the treatment of operable node-positive breast cancer, only patients with ER-positive tumours with either high Ki-67 expression or HER2 overexpression had a statistically significant improvement in disease-free survival when treated with TAC [14]. However, unlike these studies, we did not find a survival benefit from the docetaxel-containing regimen in the subset of women with HER2 positive cancer. In FinHer, half of the patients with HER2-amplified cancer were randomly assigned to receive adjuvant trastuzumab, which may have masked the potential docetaxel benefit in this subtype and may have reduced the statistical power to detect the association.
The PAM50 gene expression array has also been evaluated in predicting the potential benefit of adding a taxane to anthracycline-based chemotherapy, but none of the PAM50-derived subtypes including the luminal B subtype were predictive for a taxane benefit in the GEICAM/9966 and the NCIC CTG MA.21 randomized phase III trials [15, 16]. Similarly the Endopredict gene expression assay did not predict taxane benefit in the GEICAM/9966 study population [17].
The limitations of the study include the retrospective nature of the study, although we determined tumour MKI67 mRNA without knowledge of the clinical data and planned the statistical analyses prospectively. We tested the methods within the context of a relatively large randomized trial but lacked a validation series, and some subgroup analyses have limited power. However, the PCR method used turned out to be reproducible across multiple testing sites for all four biomarkers including MKI67 mRNA (Laible et al., manuscript submitted for publication). The details of the IHC methods used for assaying Ki-67 in the local pathology laboratories were not captured during the FinHer trial, as Ki-67 was not a protocol-mandated assay, but most pathology laboratories in Finland assess Ki-67 from the tumour hot spot areas. The recommended cut-off for ER and PgR positivity is now 1 % and no longer 10 % as it was at the time when the FinHer trial accrued patients, but the proportion of breast cancers where ER or PgR are expressed in 1 % to 10 % of nuclei is small [18].
Conclusions
Measuring of cancer ESR1, PGR and ERBB2 mRNA correlated well with the results obtained with IHC and CISH in clinical pathology laboratories. Tumour MKI67 mRNA content quantitated with RT-qPCR is associated with DDFS and OS of patients treated with modern adjuvant regimens. The results suggest that assessment of tumour MKI67 mRNA content may be valuable for selection of patients for docetaxel-containing adjuvant therapy. Since the immunohistochemical assay results for Ki-67 expression are challenging to transfer between laboratories, and the assay for measuring cancer MKI67 mRNA content with RT-qPCR might be less challenging to standardize than IHC stainings, performing studies that evaluate interlaboratory comparisons of cancer ESR1, PGR, ERBB2 and MKI67 mRNA content using RT-qPCR are warranted.
Abbreviations
- ESR1/ER:
-
Oestrogen Receptor alpha
- CISH:
-
Chromogenic in situ hybridization
- Cq:
-
Quantification cycle
- DDFS:
-
Distant disease-free survival
- FEC:
-
Fluorouracil & epirubicin, cyclophosphamide chemotherapy
- FFPE:
-
Formalin fixed paraffin embedded
- GOI:
-
Gene of interest
- HR:
-
Hazard ratio
- IHC:
-
Immunohistochemistry
- mRNA:
-
Messenger ribonucleic acid
- MKI67/Ki-67:
-
Marker of proliferation Ki-67
- OS:
-
Overall survival
- PGR/PgR:
-
Progesterone receptor
- REF:
-
Reference gene
- RT-qPCR:
-
Reverse transcription quantitative real-time polymerase chain reaction
- ERBB2/HER2:
-
Human epidermal growth factor receptor 2
References
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B et al (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 24:2206–2223
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S et al (2010) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med 134:907–922
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ et al (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 25:118–145
Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG et al (2013) An international Ki67 reproducibility study. J Natl Cancer Inst 105:1897–1906
Varga Z, Diebold J, Dommann-Scherrer C, Frick H, Kaup D, Noske A et al (2012) How reliable is Ki-67 immunohistochemistry in grade 2 breast carcinomas? A QA study of the Swiss Working Group of Breast- and Gynecopathologists. PLoS One 7:e37379
Joensuu H, Bono P, Kataja V, Alanko T, Kokko R, Asola R et al (2009) Fluorouracil, epirubicin, and cyclophosphamide with either docetaxel or vinorelbine, with or without trastuzumab, as adjuvant treatments of breast cancer: final results of the FinHer Trial. J Clin Oncol 27:5685–5692
Joensuu H, Kellokumpu-Lehtinen PL, Bono P, Alanko T, Kataja V, Asola R et al (2006) Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med 354:809–820
Kotoula V, Kalogeras KT, Kouvatseas G, Televantou D, Kronenwett R, Wirtz RM, Fountzilas G (2013) Sample parameters affecting the clinical relevance of RNA biomarkers in translational breast cancer research. Virchows Arch 462(2):141–154
Wirtz RM, Rau T, Laible M, Schlombs K, Lakis S, Wachter D, Sahin U, Hartmann A (2015) Low influence of tumor cell content on mRNA expression levels of ERBB2, ESR1, PGR and KI67 when performing the MammaTyper® RT-PCR kit. Cancer Res. doi:10.1158/1538-7445.SABCS14-P5-10-13
Milde-Langosch K, Karn T, Müller V, Witzel I, Rody A, Schmidt M, Wirtz RM (2013) Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res Treat 137(1):57–67
Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J (2010) ImmunoRatio:a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res 12(4):R56
Nitz U, Gluz O, Huober J, Kreipe HH, Kates RE, Harmann A et al (2014) Final analysis of the prospective WSG-AGO EC-Doc versus FEC phase III trial in intermediate-risk (pN1) early breast cancer: efficacy and predictive value of Ki67 expression. Ann Oncol 25:1551–1557
Jacquemier J, Boher JM, Roche H, Esterni B, Serin D, Kerbrat P et al (2011) Protein expression, survival and docetaxel benefit in node-positive breast cancer treated with adjuvant chemotherapy in the FNCLCC-PACS 01 randomized trial. Breast Cancer Res 13:R109
Hugh J, Hanson J, Cheang MC, Nielsen TO, Perou CM, Dumontet C et al (2009) Breast cancer subtypes and response to docetaxel in node-positive breast cancer: use of an immunohistochemical definition in the BCIRG 001 trial. J Clin Oncol 27:1168–1176
Liu S, Chapman JA, Burnell MJ, Levine MN, Pritchard KI, Whelan TJ et al (2015) Prognostic and predictive investigation of PAM50 intrinsic subtypes in the NCIC CTG MA.21 phase III chemotherapy trial. Breast Cancer Res Treat 149:435–448
Martin M, Prat A, Rodriquez-Lescure A, Caballero R, Ebbert MT, Munárriz B et al (2013) PAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancer. Breast Cancer Res Treat 138:457–466
Martin M, Brase JC, Calvo L, Krappmann K, Ruiz-Borrego M, Fisch K et al (2014) Clinical validation of the EndoPredict test in node-positive, chemotherapy-treated ER+/HER2- breast cancer patients: results from the GEICAM 9906 trial. Breast Cancer Res 16(2):R38
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S et al (2010) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 28:2784–2795
Acknowledgments
We thank Elke Veltrup, Susanne Scharff, Silke Claas and Torsten Acht for excellent technical support in developing molecular subtyping technologies, and. Drs. Thomas Keller and Stefan Weber for performing statistical analyses. The study was supported financially by the Academy of Finland, Cancer Society of Finland, Jane and Aatos Erkko Foundation, the Sigrid Juselius Foundation, and the Research Funds of the Helsinki University Hospital.
Author contributions
HJ, RMW and US were involved in the conception of the study. RMW performed the RT-qPCR assays; HS, JI, PH, P-LK-L, PA, TT-H, SJ and HJ provided study data and materials; SW performed the statistical analysis and wrote the statistical plan; RMW, HJ, SL, KS, ML, SE and US interpreted the data; RMW, SL and HJ drafted the manuscript; all authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
RM Wirtz is the founder and CEO of STRATIFYER Molecular Pathology GmbH. S Eidt has stock options in STRATIFYER Molecular Pathology GmbH. S Lakis is an employee of STRATIFYER Molecular Pathology GmbH. U Sahin is the founder and CEO of BioNTech Diagnostics GmbH. K Schlombs and M Laible are employees of BioNTech Diagnostics GmbH. No potential conflicts of interests were disclosed by the other authors.
Ethical standards
Each study participant provided signed informed consent for the clinical trial, and a separate consent to use tumour tissue for research related to the FinHer study. An Ethics Committee of the Helsinki University Hospital approved the protocol of the current study (HUS 124/13/03/02/2014, permission granted on Apr 9, 2014).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wirtz, R.M., Sihto, H., Isola, J. et al. Biological subtyping of early breast cancer: a study comparing RT-qPCR with immunohistochemistry. Breast Cancer Res Treat 157, 437–446 (2016). https://doi.org/10.1007/s10549-016-3835-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3835-7