Abstract
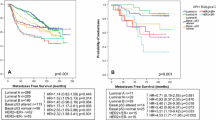
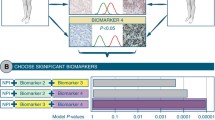
Accurate distant metastasis (DM) prediction is critical for risk stratification and effective treatment decisions in breast cancer (BC). Many prognostic markers/models based on tissue marker studies are continually emerging using conventional statistical approaches analysing complex/dimensional data association with DM/poor prognosis. However, few of them have fulfilled satisfactory evidences for clinical application. This study aimed at building DM risk assessment algorithm for BC patients. A well-characterised series of early invasive primary operable BC (n = 1902), with immunohistochemical expression of a panel of biomarkers (n = 31) formed the material of this study. Decision tree algorithm was computed using WEKA software, utilising quantitative biomarkers’ expression and the absence/presence of distant metastases. Fifteen biomarkers were significantly associated with DM, with six temporal subgroups characterised based on time to development of DM ranging from <1 to >15 years of follow-up. Of these 15 biomarkers, 10 had a significant expression pattern where Ki67LI, HER2, p53, N-cadherin, P-cadherin, PIK3CA and TOMM34 showed significantly higher expressions with earlier development of DM. In contrast, higher expressions of ER, PR and BCL2 were associated with delayed occurrence of DM. DM prediction algorithm was built utilising cases informative for the 15 significant markers. Four risk groups of patients were characterised. Three markers p53, HER2 and BCL2 predicted the probability of DM, based on software-generated cut-offs, with a precision rate of 81.1 % for positive predictive value and 77.3 %, for the negative predictive value. This algorithm reiterates the reported prognostic values of these three markers and underscores their central biological role in BC progression. Further independent validation of this pruned panel of biomarkers is therefore warranted.



Similar content being viewed by others
References
Rabbani F, Koppie TM, Charytonowicz E, Drobnjak M, Bochner BH, Cordon-Cardo C (2007) Prognostic significance of p27(Kip1) expression in bladder cancer. BJU Int 100(2):259–263
Nguyen DX, Massague J (2007) Genetic determinants of cancer metastasis. Nat Rev Genet 8(5):341–352
Pantel K, Brakenhoff RH (2004) Dissecting the metastatic cascade. Nat Rev Cancer 4(6):448–456
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2005) Reporting recommendations for tumor marker prognostic studies (REMARK). Nat Clin Pract Oncol 2(8):416–422
Cruz JA, Wishart DS (2007) Applications of machine learning in cancer prediction and prognosis. Cancer Inform 11(2):55–77
Kotsiantis SB (2007) Supervised machine learning: a review of classification techniques. Informatica 31:249–268
Salzberg S (1994) C4.5: programs for machine learning by J. Ross Quinlan. Morgan Kaufmann Publishers Inc, 1993. Mach Learn 16(3):235–240
Quinlan JR (1993) C4.5: programs for machine learning. Morgan Kaufmann Publishers, Inc., Los Altos
Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer (Journal International du Cancer) 116(3):340–350
Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet J-S, Akslen LA et al (2009) Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15(7):2302–2310
Abd El-Rehim DM, Pinder SE, Paish CE, Bell JA, Rampaul RS, Blamey RW, Robertson JF, Nicholson RI, Ellis IO (2004) Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer 91(8):1532–1542
Albasri A, Seth R, Jackson D, Benhasouna A, Crook S, Nateri AS, Chapman R, Ilyas M (2009) C-terminal Tensin-like (CTEN) is an oncogene which alters cell motility possibly through repression of E-cadherin in colorectal cancer. J Pathol 218(1):57–65
Ling LJ, Wang S, Liu XA, Shen EC, Ding Q, Lu C, Xu J, Cao QH, Zhu HQ, Wang F (2008) A novel mouse model of human breast cancer stem-like cells with high CD44 + CD24-/lower phenotype metastasis to human bone. Chin Med J (Engl) 121(20):1980–1986
Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, Robertson JFR, Aparicio S, Ellis IO, Brenton JD et al (2006) A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene 26(10):1507–1516
Zhang H, Rakha EA, Ball GR, Spiteri I, Aleskandarany M, Paish EC, Powe DG, Macmillan RD, Caldas C, Ellis IO et al (2010) The proteins FABP7 and OATP2 are associated with the basal phenotype and patient outcome in human breast cancer. Breast Cancer Res Treat 121(1):41–51
Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, Robertson JF, Aparicio S, Ellis IO, Brenton JD et al (2007) A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene 26(10):1507–1516
Aleskandarany MA, Rakha EA, Macmillan RD, Powe DG, Ellis IO, Green AR (2011) MIB1/Ki-67 labelling index can classify grade 2 breast cancer into two clinically distinct subgroups. Breast Cancer Res Treat 127(3):591–599
McCarty KS Jr, Miller LS, Cox EB, Konrath J, McCarty KS Sr (1985) Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109(8):716–721
Rakha EA, Aleskandarany M, El-Sayed ME, Blamey RW, Elston CW, Ellis IO, Lee AH (2009) The prognostic significance of inflammation and medullary histological type in invasive carcinoma of the breast. Eur J Cancer 45(10):1780–1787
Altman DG, Bland JM (1994) Statistics Notes: diagnostic tests 2: predictive values. BMJ 309(6947):102
Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schutz F et al (2008) Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res 10(4):R65
Liu Y, Kulesz-Martin MF (2006) Sliding into home: facilitated p53 search for targets by the basic DNA binding domain. Cell Death Differ 13(6):881–884
Goodsell DS (2002) The molecular perspective: Bcl-2 and apoptosis. Oncologist 7(3):259–260
Rakha EA, Putti TC, El-Rehim DMA, Paish C, Green AR, Powe DG, Lee AH, Robertson JF, Ellis IO (2006) Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol 208(4):495–506
Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14(5):1368–1376
Aleskandarany MA, Negm OH, Green AR, Ahmed MA, Nolan CC, Tighe PJ, Ellis IO, Rakha EA (2014) Epithelial mesenchymal transition in early invasive breast cancer: an immunohistochemical and reverse phase protein array study. Breast Cancer Res Treat 145(2):339–348
Aleskandarany MA, Rakha EA, Ahmed MA, Powe DG, Paish EC, Macmillan RD, Ellis IO, Green AR (2010) PIK3CA expression in invasive breast cancer: a biomarker of poor prognosis. Breast Cancer Res Treat 122(1):45–53
Zardavas D, Phillips WA, Loi S (2014) PIK3CA mutations in breast cancer: reconciling findings from preclinical and clinical data. Breast Cancer Res 16(1):201
Holmgren L, O’Reilly MS, Folkman J (1995) Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med 1(2):149–153
Townson JL, Chambers AF (2006) Dormancy of solitary metastatic cells. Cell Cycle 5(16):1744–1750
Abdel-Fatah TM, Powe DG, Agboola J, Adamowicz-Brice M, Blamey RW, Lopez-Garcia MA, Green AR, Reis-Filho JS, Ellis IO (2010) The biological, clinical and prognostic implications of p53 transcriptional pathways in breast cancers. J Pathol 220(4):419–434
Cianfrocca M, Goldstein LJ (2004) Prognostic and predictive factors in early-stage breast cancer. Oncologist 9(6):606–616
Callagy GM, Webber MJ, Pharoah PD, Caldas C (2008) Meta-analysis confirms BCL2 is an independent prognostic marker in breast cancer. BMC Cancer 8:153
Conflict of interest
The authors have no conflicts of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aleskandarany, M.A., Soria, D., Green, A.R. et al. Markers of progression in early-stage invasive breast cancer: a predictive immunohistochemical panel algorithm for distant recurrence risk stratification. Breast Cancer Res Treat 151, 325–333 (2015). https://doi.org/10.1007/s10549-015-3406-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-015-3406-3