Abstract
Anti-estrogen therapies are not effective in ER− breast cancers, thus identifying mechanisms underlying lack of ER expression in ER− breast cancers is imperative. We have previously demonstrated that hyperactivation of MAPK (hMAPK) downstream of overexpressed EGFR or overexpression/amplification of Her2 represses ER protein and mRNA expression. Abrogation of hMAPK in ER− breast cancer cell lines and primary cultures causes re-expression of ER and restoration of anti-estrogen responses. This study was performed to identify mechanisms of hMAPK-induced transcriptional repression of ER. We found that ER promoter activity is significantly reduced in the presence of hMAPK signaling, yet did not identify specific promoter sequences responsible for this repression. We performed an epigenetic compound screen in an ER− breast cancer cell line that expresses hMAPK yet does not exhibit ER promoter hypermethylation. A number of HDAC inhibitors were identified and confirmed to modulate ER expression and estrogen signaling in multiple ER− cell lines and tumor samples lacking ER promoter methylation. siRNA-mediated knockdown of HDACs 1, 2, and 3 reversed the mRNA repression in multiple breast cancer cell lines and primary cultures and ER promoter-associated histone acetylation increased following MAPK inhibition. These data implicate histone deacetylation downstream of hMAPK in the observed ER mRNA repression associated with hMAPK. Importantly, histone deacetylation appears to be a common mechanism in the transcriptional repression of ER between ER− breast cancers with or without ER promoter hypermethylation.









Similar content being viewed by others
References
Polyak K (2007) Breast cancer: origins and evolution. J Clin Invest 117(11):3155–3163
Saez RA, McGuire WL, Clark GM (1989) Prognostic factors in breast cancer. Semin Surg Oncol 5(2):102–110
Knight WA, Livingston RB, Gregory EJ, McGuire WL (1977) Estrogen receptor as an independent prognostic factor for early recurrence in breast cancer. Cancer Res 37(12):4669–4671
Sommer S, Fuqua SA (2001) Estrogen receptor and breast cancer. Semin Cancer Biol 11(5):339–352
Wright C, Angus B, Napier J, Wetherall M, Udagawa Y, Sainsbury JR, Johnston S, Carpenter F, Horne CH (1987) Prognostic factors in breast cancer: immunohistochemical staining for SP1 and NCRC 11 related to survival, tumour epidermal growth factor receptor and oestrogen receptor status. J Pathol 153(4):325–331
Allred DC, Mohsin SK, Fuqua SAW (2001) Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer 8:47–61
Lapidus RG, Ferguson AT, Ottaviano YL, Parl FF, Smith HS, Weitzman SA, Baylin SB, Issa JP, Davidson NE (1996) Methylation of estrogen and progesterone receptor gene 5′ CpG islands correlates with lack of estrogen and progesterone receptor gene expression in breast tumors. Clin Cancer Res 2(5):805–810
Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE (1994) Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res 54(10):2552–2555
Ferguson AT, Lapidus RG, Baylin SB, Davidson NE (1995) Demethylation of the estrogen receptor gene in estrogen receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res 55(11):2279–2283
Sainsbury JR, Farndon JR, Harris AL, Sherbet GV (1985) Epidermal growth factor receptors on human breast cancers. Br J Surg 72(3):186–188
Oh AS, Lorant LA, Holloway JN, Miller DL, Kern FG, El-Ashry D (2001) Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol Endocrinol 15(8):1344–1359
Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D (2006) Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res 66(7):3903–3911
Liu Y, El-Ashry D, Chen D, Ding IY, Kern FG (1995) MCF-7 breast cancer cells overexpressing transfected c-erbB-2 have an in vitro growth advantage in estrogen-depleted conditions and reduced estrogen-dependence and tamoxifen-sensitivity in vivo. Breast Cancer Res Treat 34(2):97–117
Miller DL, El-Ashry D, Cheville AL, Liu Y, McLeskey SW, Kern FG (1994) Emergence of MCF-7 cells overexpressing a transfected epidermal growth factor receptor (EGFR) under estrogen-depleted conditions: evidence for a role of EGFR in breast cancer growth and progression. Cell Growth Differ 5(12):1263–1274
El-Ashry D, Miller DL, Kharbanda S, Lippman ME, Kern FG (1997) Constitutive Raf-1 kinase activity in breast cancer cells induces both estrogen-independent growth and apoptosis. Oncogene 15(4):423–435
Holloway JN, Murthy S, El-Ashry D (2004) A cytoplasmic substrate of mitogen-activated protein kinase is responsible for estrogen receptor-alpha down-regulation in breast cancer cells: the role of nuclear factor-kappaB. Mol Endocrinol 18(6):1396–1410
Bayliss J, Hilger A, Vishnu P, Diehl K, El-Ashry D (2007) Reversal of the estrogen receptor negative phenotype in breast cancer and restoration of antiestrogen response. Clin Cancer Res 13(23):7029–7036
deConinck EC, McPherson LA, Weigel RJ (1995) Transcriptional regulation of estrogen receptor in breast carcinomas. Mol Cell Biol 15(4):2191–2196
Drews-Elger K, Brinkman JA, Miller P, Shah SH, Harrell JC, da Silva TG, Ao Z, Schlater A, Azzam DJ, Diehl K, Thomas D, Slingerland JM, Perou CM, Lippman ME, El-Ashry D (2014) Primary breast tumor-derived cellular models: characterization of tumorigenic, metastatic, and cancer-associated fibroblasts in dissociated tumor (DT) cultures. Breast Cancer Res Treat 144(3):503–517
Das PM, Ramachandran K, vanWert J, Singal R (2004) Chromatin immunoprecipitation assay. Biotechniques 37(6):961–969
Jang ER, Lim SJ, Lee ES, Jeong G, Kim TY, Bang YJ, Lee JS (2004) The histone deacetylase inhibitor trichostatin A sensitizes estrogen receptor alpha-negative breast cancer cells to tamoxifen. Oncogene 23(9):1724–1736
Sharma D, Blum J, Yang X, Beaulieu N, Macleod AR, Davidson NE (2005) Release of methyl CpG binding proteins and histone deacetylase 1 from the estrogen receptor alpha (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol Endocrinol 19(7):1740–1751
Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, Davidson NE (2001) Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res 61(19):7025–7029
Yi X, Wei W, Wang SY, Du ZY, Xu YJ, Yu XD (2008) Histone deacetylase inhibitor SAHA induces ERalpha degradation in breast cancer MCF-7 cells by CHIP-mediated ubiquitin pathway and inhibits survival signaling. Biochem Pharmacol 75(9):1697–1705
Alao JP, Lam EW, Ali S, Buluwela L, Bordogna W, Lockey P, Varshochi R, Stavropoulou AV, Coombes RC, Vigushin DM (2004) Histone deacetylase inhibitor trichostatin A represses estrogen receptor alpha-dependent transcription and promotes proteasomal degradation of cyclin D1 in human breast carcinoma cell lines. Clin Cancer Res 10(23):8094–8104
Stossi F, Madak-Erdogan Z, Katzenellenbogen BS (2012) Macrophage-elicited loss of estrogen receptor-alpha in breast cancer cells via involvement of MAPK and c-Jun at the ESR1 genomic locus. Oncogene 31(14):1825–1834
Yang SH, Vickers E, Brehm A, Kouzarides T, Sharrocks AD (2001) Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol Cell Biol 21(8):2802–2814
Ferguson BS, Harrison BC, Jeong MY, Reid BG, Wempe MF, Wagner FF, Holson EB, McKinsey TA (2013) Signal-dependent repression of DUSP5 by class I HDACs controls nuclear ERK activity and cardiomyocyte hypertrophy. Proc Natl Acad Sci USA 110(24):9806–9811
Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, Boesche M, Delling M, Dumpelfeld B, Eberhard D, Huthmacher C, Mathieson T, Poeckel D, Reader V, Strunk K, Sweetman G, Kruse U, Neubauer G, Ramsden NG, Drewes G (2011) Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol 29(3):255–265
Williams KA, Zhang M, Xiang S, Hu C, Wu JY, Zhang S, Ryan M, Cox AD, Der CJ, Fang B, Koomen J, Haura E, Bepler G, Nicosia SV, Matthias P, Wang C, Bai W, Zhang X (2013) Extracellular signal-regulated kinase (ERK) phosphorylates histone deacetylase 6 (HDAC6) at serine 1035 to stimulate cell migration. J Biol Chem 288(46):33156–33170
Zhou X, Richon VM, Wang AH, Yang XJ, Rifkind RA, Marks PA (2000) Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc Natl Acad Sci USA 97(26):14329–14333
Krusche CA, Wulfing P, Kersting C, Vloet A, Bocker W, Kiesel L, Beier HM, Alfer J (2005) Histone deacetylase-1 and -3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res Treat 90(1):15–23
Muller B, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W, Denkert C (2013) Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer—overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer 13:215
Kawai H, Li H, Avraham S, Jiang S, Avraham HK (2003) Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int J Cancer 107(3):353–358
Winter M, Moser MA, Meunier D, Fischer C, Machat G, Mattes K, Lichtenberger BM, Brunmeir R, Weissmann S, Murko C, Humer C, Meischel T, Brosch G, Matthias P, Sibilia M, Seiser C (2013) Divergent roles of HDAC1 and HDAC2 in the regulation of epidermal development and tumorigenesis. EMBO J 32(24):3176–3191
Brinkman JA, El-Ashry D (2009) ER re-expression and re-sensitization to endocrine therapies in ER-negative breast cancers. J Mammary Gland Biol Neoplasia 14(1):67–78
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH 1R01 CA113674 to DEA and NIH 1R01NS067289 to NA), the Bankhead Coley Foundation (BC-09BW-04-RC1 to DEA), and by the Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine (to DEA). We would like to thank the Center for Therapeutic Innovation at the University of Miami Miller School of Medicine for the use of their epigenetic compound library and laboratory equipment, and Laura Parsons and Joeli Brinkman for technical contributions. We would also like to thank Drs. Marc Lippman, Zafar Nawaz, and Kerry Burnstein of the University of Miami for thoughtful discussion.
Disclosures
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
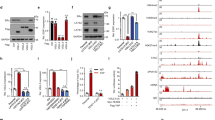
Graph shows ESR1 promoter methylation status in a number of primary tumor cell lines, SUM cell lines, and engineered hyperactive ERK1/2 MAP kinase MCF-7 cells analyzed by the Epitect Methyl-II PCR assay (Qiagen). MDA-MB-231 serves as a positive control for ESR1 promoter methylation while MCF-7 cells are the negative control. Red boxes identify those cell lines utilized for screening and validation studies. Supplementary material 1 (PDF 33 kb)
Online Resource 2
a Plasmid map of SABiosciences pGreenFire lentiviral reporter vector. 4 tandem EREs were inserted into the multiple cloning site prior to the minimal CMV promoter upstream of GFP and luciferase genes. In the absence of ER activation, there is no expression of GFP or luciferase. b 24 h treatment with ethanol vehicle, 10 nM estradiol, 100 nM faslodex, or estradiol + faslodex. Y-axis is relative luciferase units (RLU). **p < 0.001. c Graphic representation of luciferase expression in response to compound + estradiol treatment. Expression is shown as percent of baseline expression, as calculated from untreated wells of the compound plate. Three standard deviations from the mean gives a range of 71–129 % of baseline leading to a number of candidates providing a statistically significant increase in luciferase expression, the highest expressers being HDAC inhibitors. Supplementary material 2 (PDF 567 kb)
Online Resource 3
Compounds represented in the plate-based compound screen. Compounds include: HDAC inhibitors, HDAC stimulators, histone lysine deacetylase inhibitors, HIF1a inhibitor, histone demethylase, histone acetyltransferase inhibitor, sirtuin inhibitors (type III HDAC), etc. Supplementary material 3 (PDF 47 kb)
Online Resource 4
Feedback loop involving class I HDACs, ERK1/2, and DUSP5. We hypothesize that class I HDAC inhibition leading to increased DUSP5 expression leads to reduced ERK1/2 phosphorylation/activation, leading to further decreased class I HDAC activity. This reduced HDAC activity further increases both DUSP5 and ER expression, leading to further reduced ERK1/2 phosphorylation. Supplementary material 4 (PDF 546 kb)
Rights and permissions
About this article
Cite this article
Plotkin, A., Volmar, CH., Wahlestedt, C. et al. Transcriptional repression of ER through hMAPK dependent histone deacetylation by class I HDACs. Breast Cancer Res Treat 147, 249–263 (2014). https://doi.org/10.1007/s10549-014-3093-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-3093-5