Abstract
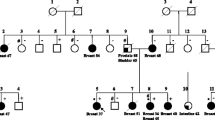
Bloom’s syndrome is a rare autosomal recessive chromosomal instability disorder with a high incidence of various types of neoplasia, including breast cancer. Whether monoallelic BLM mutations predispose to breast cancer has been a long-standing question. A nonsense mutation, p.Q548X, has recently been associated with an increased risk for breast cancer in a Russian case–control study. In the present work, we have investigated the prevalence of this Slavic BLM founder mutation in a total of 3,188 breast cancer cases and 2,458 controls from Bashkortostan, Belarus, Ukraine, and Kazakhstan. The p.Q548X allele was most frequent in Russian patients (0.8 %) but was also prevalent in Byelorussian and Ukrainian patients (0.5 and 0.6 %, respectively), whereas it was absent in Altaic or other non-European subpopulations. In a combined analysis of our four case–control series, the p.Q548X mutation was significantly associated with breast cancer (Mantel–Haenszel OR 5.1, 95 % CI 1.2; 21.9, p = 0.03). A meta-analysis with the previous study from the St. Petersburg area corroborates the association (OR 5.7, 95 % CI 2.0; 15.9, p = 3.7 × 10−4). A meta-analysis for all published truncating mutations further supports the association of BLM with breast cancer, with an estimated two- to five-fold increase in risk (OR 3.3, 95 %CI 1.9; 5.6, p = 1.9 × 10−5). Altogether, these data indicate that BLM is not only a gene for Bloom’s syndrome but also might represent a breast cancer susceptibility gene.
Similar content being viewed by others
References
Turnbull C, Rahman N (2008) Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet 9:321–345
Campeau PM, Foulkes WD, Tischkowitz MD (2008) Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet 124:31–42
Bogdanova N, Dörk T (2012) Molecular genetics of breast and ovarian cancer. Balkan J Med Genet 15(1):75–80
Ellis NA, German J (1996) Molecular genetics of Bloom’s syndrome. Hum Mol Genet 5:1457–1463
German J (1993) Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine 72:393–406
German J, Archibald R, Bloom D (1965) Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science 148:506–507
Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M et al (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 83:655–666
Ababou M, Dutertre S, Lécluse Y, Onclercq R, Chatton B, Amor-Guéret M (2000) ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene 19:5955–5963
Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J (2001) Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol 153:367–380
Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF (2002) Functional link between BLM defective in Bloom’s syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem 277:30515–30523
Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14:927–939
Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC (2011) BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25:350–362
Wu L, Davies SL, Levitt NC, Hickson ID (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem 276:19375–19381
Braybrooke JP, Li JL, Wu L, Caple F, Benson FE, Hickson ID (2003) Functional interaction between the Bloom’s syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D). J Biol Chem 278:48357–48366
Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA (2009) SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol 7(12):e1000252
Suhasini AN, Brosh RM Jr (2012) Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ–BLM helicase interaction. Trends Genet 28:7–13
Chan KL, North PS, Hickson ID (2007) BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 26(14):3397–3409
German J, Bloom D, Passarge E, Fried K, Goodman RM, Katzenellenbogen I, Laron Z et al (1977) Bloom’s syndrome. VI. The disorder in Israel and an estimation of the gene frequency in the Ashkenazim. Am J Hum Genet 29:553–562
Gruber SB, Ellis NA, Scott KK, Almog R, Kolachana P, Bonner JD, Kirchhoff T, Tomsho LP, Nafa K, Pierce H, Low M, Satagopan J, Rennert H, Huang H, Greenson JK, Groden J, Rapaport B, Shia J, Johnson S, Gregersen PK, Harris CC, Boyd J, Rennert G, Offit K (2002) BLM heterozygosity and the risk of colorectal cancer. Science 297:2013
Cleary SP, Zhang W, Di Nicola N, Aronson M, Aube J, Steinman A, Haddad R, Redston M, Gallinger S, Narod SA, Gryfe R (2003) Heterozygosity for the BLM(Ash) mutation and cancer risk. Cancer Res 63:1769–1771
Zauber NP, Sabbath-Solitare M, Marotta S, Zauber AG, Foulkes W, Chan M, Turner F, Bishop DT (2005) Clinical and genetic findings in an Ashkenazi Jewish population with colorectal neoplasms. Cancer 104:719–729
Sokolenko AP, Iyevleva AG, Preobrazhenskaya EV, Mitiushkina NV, Abysheva SN, Suspitsin EN, Kuligina ESh, Gorodnova TV, Pfeifer W, Togo AV, Turkevich EA, Ivantsov AO, Voskresenskiy DV, Dolmatov GD, Bit-Sava EM, Matsko DE, Semiglazov VF, Fichtner I, Larionov AA, Kuznetsov SG, Antoniou AC, Imyanitov EN (2012) High prevalence and breast cancer predisposing role of the BLM c.1642 C > T (Q548X) mutation in Russia. Int J Cancer 130:2867–2873
Bogdanova N, Cybulski C, Bermisheva M, Datsyuk I, Yamini P, Hillemanns P, Antonenkova NN, Khusnutdinova E, Lubinski J, Dörk T (2009) A nonsense mutation (E1978X) in the ATM gene is associated with breast cancer. Breast Cancer Res Treat 118:207–211
Bogdanova NV, Antonenkova NN, Rogov YI, Karstens JH, Hillemanns P, Dörk T (2010) High frequency and allele-specific differences of BRCA1 founder mutations in breast cancer and ovarian cancer patients from Belarus. Clin Genet 78:364–372
Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, Dennis J et al (2012) Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet 44:312–318
Bogdanova N, Feshchenko S, Cybulski C, Dörk T (2007) CHEK2 mutation and hereditary breast cancer. J Clin Oncol 25(19):e26
Koren-Michowitz M, Friedman E, Gershoni-Baruch R, Brok-Simoni F, Patael Y, Rechavi G, Amariglio N (2005) Coinheritance of BRCA1 and BRCA2 mutations with Fanconi anemia and Bloom syndrome mutations in Ashkenazi Jewish population: possible role in risk modification for cancer development. Am J Hematol 78:203–206
Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DY, Tothill RW, Thorne H, kConFab, Barnes DR, Li J, Ellul J, Philip GK, Antill YC, James PA, Trainer AH, Mitchell G, Campbell IG (2012) Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet 8(9):e1002894
German J (1997) Bloom’s syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet 93:100–106
Masmoudi A, Marrakchi S, Kamoun H, Chaaben H, Ben Salah G, Ben Salah R, Fakhfakh F, Zahaf A, Turki H (2012) Clinical and laboratory findings in 8 patients with Bloom’s syndrome. J Dermatol Case Rep 6:29–33
Baris HN, Kedar I, Halpern GJ, Shohat T, Magal N, Ludman MD, Shohat M (2007) Prevalence of breast and colorectal cancer in Ashkenazi Jewish carriers of Fanconi anemia and Bloom syndrome. Isr Med Assoc J 9:847–850
Ellis NA, Offit K (2012) Heterozygous mutations in DNA repair genes and hereditary breast cancer: a question of power. PLoS Genet 8(9):e1003008
Górski B, Cybulski C, Huzarski T, Byrski T, Gronwald J, Jakubowska A, Stawicka M, Gozdecka-Grodecka S, Szwiec M, Urbański K, Mituś J, Marczyk E, Dziuba J, Wandzel P, Surdyka D, Haus O, Janiszewska H, Debniak T, Tołoczko-Grabarek A, Medrek K, Masojć B, Mierzejewski M, Kowalska E, Narod SA, Lubiński J (2005) Breast cancer predisposing alleles in Poland. Breast Cancer Res Treat 92:19–24
Bogdanova N, Feshchenko S, Schürmann P, Waltes R, Wieland B, Hillemanns P, Rogov YI, Dammann O, Bremer M, Karstens JH, Sohn C, Varon R, Dörk T (2008) Nijmegen Breakage Syndrome mutations and risk of breast cancer. Int J Cancer 122:802–806
Heidemann S, Fischer C, Engel C, Fischer B, Harder L, Schlegelberger B, Niederacher D, Goecke TO, Doelken SC, Dikow N, Jonat W, Morlot S, Schmutzler RC, Arnold NK (2012) Double heterozygosity for mutations in BRCA1 and BRCA2 in German breast cancer patients: implications on test strategies and clinical management. Breast Cancer Res Treat 134:1229–1239
Pern F, Bogdanova N, Schürmann P, Lin M, Ay A, Länger F, Hillemanns P, Christiansen H, Park-Simon TW, Dörk T (2012) Mutation analysis of BRCA1, BRCA2, PALB2 and BRD7 in a hospital-based series of German patients with triple-negative breast cancer. PLoS One 7(10):e47993
Turnbull C, Seal S, Renwick A, Warren-Perry M, Hughes D, Elliott A, Pernet D, Peock S, Adlard JW, Barwell J, Berg J, Brady AF, Brewer C, Brice G, Chapman C, Cook J, Davidson R, Donaldson A, Douglas F, Greenhalgh L, Henderson A, Izatt L, Kumar A, Lalloo F, Miedzybrodzka Z, Morrison PJ, Paterson J, Porteous M, Rogers MT, Shanley S, Walker L, Breast Cancer Susceptibility Collaboration (UK), EMBRACE, Ahmed M, Eccles D, Evans DG, Donnelly P, Easton DF, Stratton MR, Rahman N (2012) Gene-gene interactions in breast cancer susceptibility. Hum Mol Genet 21:958–962
Hoadley KA, Xue Y, Ling C, Takata M, Wang W, Keck JL (2012) Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome. Proc Natl Acad Sci USA 109:4437–4442
Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet 26:424–429
Acknowledgments
We thank the patients and healthy volunteers for their participation and the clinicians at hospitals in Belarus, Kazakhstan, Russia, and Ukraine for their support of this work. We cordially thank Evgeny Imyanitov and his group for providing us with a positive control for this study. D.P. was supported by travel funds from the German Ministry of Education and Research and by grant 12-04-97026 of the Russian Foundation for Basic Research. N.B. was supported by an intramural Hannelore-Munke stipend at Hannover Medical School. I.D. was supported by a German Academic Exchange Program (DAAD). The Hannover laboratory was furthermore supported by the Rudolf Bartling Foundation.
Ethical Standards
The experiments in the present study comply with the current laws of the country in which they were performed.
Conflict of interest
None of the authors declares a conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2012_2357_MOESM1_ESM.tif
Supplementary material 1 (TIFF 269 kb): Identification of the BLM mutation p.Q548X by means of high-resolution melting analysis (top) and direct sequencing (bottom). Top: Normalized fluorescence graph for high-resolution melting analysis of the mutant amplicon carrying the p.Q548X mutation in comparison with wildtype controls. Bottom: Direct sequencing of exon 7 in the BLM gene, sense strand, in a sample with heterozygosity for mutation p.Q548X (C/T, asterisk)
Rights and permissions
About this article
Cite this article
Prokofyeva, D., Bogdanova, N., Dubrowinskaja, N. et al. Nonsense mutation p.Q548X in BLM, the gene mutated in Bloom’s syndrome, is associated with breast cancer in Slavic populations. Breast Cancer Res Treat 137, 533–539 (2013). https://doi.org/10.1007/s10549-012-2357-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-012-2357-1