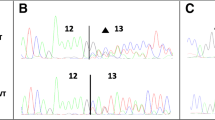
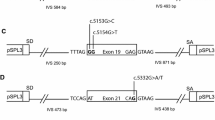
Abstract
Mutations in BRCA1 and BRCA2 predispose carriers to early onset breast and ovarian cancer. A common problem in clinical genetic testing is interpretation of variants with unknown clinical significance. The Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium was initiated to evaluate and implement strategies to characterize the clinical significance of BRCA1 and BRCA2 variants. As an initial project of the ENIGMA Splicing Working Group, we report splicing and multifactorial likelihood analysis of 25 BRCA1 and BRCA2 variants from seven different laboratories. Splicing analysis was performed by reverse transcriptase PCR or mini gene assay, and sequencing to identify aberrant transcripts. The findings were compared to bioinformatic predictions using four programs. The posterior probability of pathogenicity was estimated using multifactorial likelihood analysis, including co-occurrence with a deleterious mutation, segregation and/or report of family history. Abnormal splicing patterns expected to lead to a non-functional protein were observed for 7 variants (BRCA1 c.441+2T>A, c.4184_4185+2del, c.4357+1G>A, c.4987-2A>G, c.5074G>C, BRCA2 c.316+5G>A, and c.8754+3G>C). Combined interpretation of splicing and multifactorial analysis classified an initiation codon variant (BRCA2 c.3G>A) as likely pathogenic, uncertain clinical significance for 7 variants, and indicated low clinical significance or unlikely pathogenicity for another 10 variants. Bioinformatic tools predicted disruption of consensus donor or acceptor sites with high sensitivity, but cryptic site usage was predicted with low specificity, supporting the value of RNA-based assays. The findings also provide further evidence that clinical RNA-based assays should be extended from analysis of invariant dinucleotides to routinely include all variants located within the donor and acceptor consensus splicing sites. Importantly, this study demonstrates the added value of collaboration between laboratories, and across disciplines, to collate and interpret information from clinical testing laboratories to consolidate patient management.


Similar content being viewed by others
References
Thomassen M, Hansen TV, Borg A, Lianee HT, Wikman F, Pedersen IS, Bisgaard ML, Nielsen FC, Kruse TA, Gerdes AM (2008) BRCA1 and BRCA2 mutations in Danish families with hereditary breast and/or ovarian cancer. Acta Oncol 47:772–777
Diez O, Osorio A, Duran M, Martinez-Ferrandis JI, de la Hoya M, Salazar R, Vega A, Campos B, Rodriguez-Lopez R, Velasco E, Chaves J, Diaz-Rubio E, Jesus CJ, Torres M, Esteban E, Cervantes A, Alonso C, San Roman JM, Gonzalez-Sarmiento R, Miner C, Carracedo A, Eugenia AM, Caldes T, Benitez J, Baiget M (2003) Analysis of BRCA1 and BRCA2 genes in Spanish breast/ovarian cancer patients: a high proportion of mutations unique to Spain and evidence of founder effects. Hum Mutat 22:301–312
Malacrida S, Agata S, Callegaro M, Casella C, Barana D, Scaini MC, Manoukian S, Oliani C, Radice P, Barile M, Menin C, D’Andrea E, Montagna M (2008) BRCA1 p.Val1688del is a deleterious mutation that recurs in breast and ovarian cancer families from Northeast Italy. J Clin Oncol 26:26–31
Zhang MQ (1998) Statistical features of human exons and their flanking regions. Hum Mol Genet 7:919–932
Yeo G, Burge CB (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394
Reese MG, Eeckman FH, Kulp D, Haussler D (1997) Improved splice site detection in Genie. J Comput Biol 4:311–323
Pertea M, Lin X, Salzberg SL (2001) GeneSplicer: a new computational method for splice site prediction. Nucl Acids Res 29:1185–1190
Fairbrother WG, Yeh RF, Sharp PA, Burge CB (2002) Predictive identification of exonic splicing enhancers in human genes. Science 297:1007–1013
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucl Acids Res 31:3568–3571
Spurdle AB, Couch FJ, Hogervorst FB, Radice P, Sinilnikova OM (2008) Prediction and assessment of splicing alterations: implications for clinical testing. Hum Mutat 29:1304–1313
Vreeswijk MP, Kraan JN, van der Klift HM, Vink GR, Cornelisse CJ, Wijnen JT, Bakker E, van Asperen CJ, Devilee P (2009) Intronic variants in BRCA1 and BRCA2 that affect RNA splicing can be reliably selected by splice-site prediction programs. Hum Mutat 30:107–114
Walker LC, Whiley PJ, Couch FJ, Farrugia DJ, Healey S, Eccles DM, Lin F, Butler SA, Goff SA, Thompson BA, Lakhani SR, Da Silva LM, Tavtigian SV, Goldgar DE, Brown MA, Spurdle AB (2010) Detection of splicing aberrations caused by BRCA1 and BRCA2 sequence variants encoding missense substitutions: implications for prediction of pathogenicity. Hum Mutat 31:E1484–E1505
Tesoriero AA, Wong EM, Jenkins MA, Hopper JL, Brown MA, Chenevix-Trench G, Spurdle AB, Southey MC (2005) Molecular characterization and cancer risk associated with BRCA1 and BRCA2 splice site variants identified in multiple-case breast cancer families. Hum Mutat 26:495
Spurdle AB, Lakhani SR, Da Silva LM, Balleine RL, Goldgar DE (2010) Bayes analysis provides evidence of pathogenicity for the BRCA1 c.135-1G>T (IVS3-1) and BRCA2 c.7977–1G>C (IVS17-1) variants displaying in vitro splicing results of equivocal clinical significance. Hum Mutat 31:E1141–E1145
Santos C, Peixoto A, Rocha P, Vega A, Soares MJ, Cerveira N, Bizarro S, Pinheiro M, Pereira D, Rodrigues H, Castro F, Henrique R, Teixeira MR (2009) Haplotype and quantitative transcript analyses of Portuguese breast/ovarian cancer families with the BRCA1 R71G founder mutation of Galician origin. Fam Cancer 8:203–208
Bonnet C, Krieger S, Vezain M, Rousselin A, Tournier I, Martins A, Berthet P, Chevrier A, Dugast C, Layet V, Rossi A, Lidereau R, Frebourg T, Hardouin A, Tosi M (2008) Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet 45:438–446
Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ (2004) Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet 75:535–544
Couch FJ, Rasmussen LJ, Hofstra R, Monteiro AN, Greenblatt MS, de WN (2008) Assessment of functional effects of unclassified genetic variants. Hum Mutat 29:1314–1326
Hansen TV, Steffensen AY, Jonson L, Andersen MK, Ejlertsen B, Nielsen FC (2010) The silent mutation nucleotide 744 G→A, Lys172Lys, in exon 6 of BRCA2 results in exon skipping. Breast Cancer Res Treat 119:547–550
Mohammadi L, Vreeswijk MP, Oldenburg R, van den Ouweland A, Oosterwijk JC, van der Hout AH, Hoogerbrugge N, Ligtenberg M, Ausems MG, van der Luijt RB, Dommering CJ, Gille JJ, Verhoef S, Hogervorst FB, van Os TA, Gomez GE, Blok MJ, Wijnen JT, Helmer Q, Devilee P, van Asperen CJ, van Houwelingen HC (2009) A simple method for co-segregation analysis to evaluate the pathogenicity of unclassified variants; BRCA1 and BRCA2 as an example. BMC Cancer 9:211
Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, Goldgar DE (2007) A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet 81:873–883
Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV (2008) Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 29:1282–1291
Santarosa M, Viel A, Boiocchi M (1999) Splice variant lacking the transactivation domain of the BRCA2 gene and mutations in the splice acceptor site of intron 2. Genes Chromosomes Cancer 26:381–382
Diez O, Gutierrez-Enriquez S, Cajal T, Alonso C, Balmana J, Llort G (2007) Caution should be used when interpreting alterations affecting the exon 3 of the BRCA2 gene in breast/ovarian cancer families. J Clin Oncol 25:5035–5036
Coupier I, Baldeyron C, Rousseau A, Mosseri V, Pages-Berhouet S, Caux-Moncoutier V, Papadopoulo D, Stoppa-Lyonnet D (2004) Fidelity of DNA double-strand break repair in heterozygous cell lines harbouring BRCA1 missense mutations. Oncogene 23:914–919
Brandao RD, van RK, Tserpelis D, Garcia EG, Blok MJ (2011) Characterisation of unclassified variants in the BRCA1/2 genes with a putative effect on splicing. Breast Cancer Res Treat. doi:10.1007/s10549-011-1599-7
Hansen TV, Bisgaard ML, Jonson L, Albrechtsen A, Filtenborg-Barnkob B, Eiberg H, Ejlertsen B, Nielsen FC (2008) Novel de novo BRCA2 mutation in a patient with a family history of breast cancer. BMC Med Genet 9:58
Nordling M, Karlsson P, Wahlstrom J, Engwall Y, Wallgren A, Martinsson T (1998) A large deletion disrupts the exon 3 transcription activation domain of the BRCA2 gene in a breast/ovarian cancer family. Cancer Res 58:1372–1375
Sanz DJ, Acedo A, Infante M, Duran M, Perez-Cabornero L, Esteban-Cardenosa E, Lastra E, Pagani F, Miner C, Velasco EA (2010) A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin Cancer Res 16:1957–1967
Peixoto A, Santos C, Pinheiro M, Pinto P, Soares MJ, Rocha P, Gusmao L, Amorim A, van der Hout A, Gerdes AM, Thomassen M, Kruse TA, Cruger D, Sunde L, Bignon YJ, Uhrhammer N, Cornil L, Rouleau E, Lidereau R, Yannoukakos D, Pertesi M, Narod S, Royer R, Costa MM, Lazaro C, Feliubadalo L, Grana B, Blanco I, de la Hoya M, Caldes T, Maillet P, Benais-Pont G, Pardo B, Laitman Y, Friedman E, Velasco EA, Duran M, Miramar MD, Valle AR, Calvo MT, Vega A, Blanco A, Diez O, Gutierrez-Enriquez S, Balmana J, Ramon YC, Alonso C, Baiget M, Foulkes W, Tischkowitz M, Kyle R, Sabbaghian N, Ashton-Prolla P, Ewald IP, Rajkumar T, Mota-Vieira L, Giannini G, Gulino A, Achatz MI, Carraro DM, de Paillerets BB, Remenieras A, Benson C, Casadei S, King MC, Teugels E, Teixeira MR (2010) International distribution and age estimation of the Portuguese BRCA2 c.156_157insAlu founder mutation. Breast Cancer Res Treat 127(3):671–679
Machado PM, Brandao RD, Cavaco BM, Eugenio J, Bento S, Nave M, Rodrigues P, Fernandes A, Vaz F (2007) Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol 25:2027–2034
Acknowledgments
We thank the families participating in this research, the clinical personnel involved in aspects of recruitment and clinical data collection, and the clinical and research institutions supporting the combined research efforts. Melissa Brown is thanked for collecting the protocols used by the different laboratories. FPGMX: This work was partially supported by grants from the Xunta de Galicia (10PXIB 9101297PR) and FMM Foundation given to A.V. L.F is supported by Isabel Barreto program from Xunta de Galicia and Fondo Social Europeo. ICO: Contract grant sponsor: Spanish Health Research Fund; Carlos III Health Institute; Catalan Health Institute and Autonomous Government of Catalonia. Contract grant numbers: ISCIIIRETIC RD06/0020/1051, PI10/01422 and 2009SGR290. HVH: This work was partially funded by two grants (OD, 2008; SGE, 2008) from Fundación de Investigación Médica Mutua Madrileña. CBCS: We would like to thank the NEYE foundation and Familien Hede Nielsens fond for financial support. ABS is supported by an NHMRC Senior Research Fellowship, and her research on BRCA1/2 variants is funded by an NHMRC project grant. IOV: This study was supported by “Ministero della Salute” (grant numbers RFPS 2006-5-341353, ACC2/R6.9 and “Progetto Tumori Femminili”)
Conflicts of interest
All authors declared no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Amanda B. Spurdle and Ana Vega contributing equally to this study.
This study is conducted on behalf of the ENIGMA Consortium Splicing Working Group.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thomassen, M., Blanco, A., Montagna, M. et al. Characterization of BRCA1 and BRCA2 splicing variants: a collaborative report by ENIGMA consortium members. Breast Cancer Res Treat 132, 1009–1023 (2012). https://doi.org/10.1007/s10549-011-1674-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1674-0