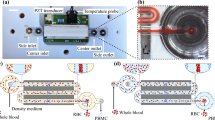
Abstract
Blood is a valuable tissue containing cellular populations rich in information regarding the immediate immune and inflammatory status of the body. Blood leukocytes or white blood cells (WBCs) provide an ideal sample to monitor systemic changes and understand molecular signaling mechanisms in disease processes. Blood samples need to be processed to deplete contaminating erythrocytes or red blood cells (RBCs) and sorted into different WBC sub-populations prior to analysis. This is typically accomplished using immuno-affinity protocols which result in undesirable activation. An alternative is size based sorting which by itself is unsuitable for WBCs sorting due to size overlap between different sub-populations. To overcome this limitation, we investigated the possibility of using controlled osmotic exposure to deplete and/or create a differential size increase between WBC populations. Using a new microfluidic cell docking platform, the response of RBCs and WBCs to deionized (DI) water was evaluated. Time lapse microscopy confirms depletion of RBCs within 15 s and creation of > 3 μm size difference between lymphocytes, monocytes and granulocytes. A flow through microfluidic device was also used to expose different WBCs to DI water for 30, 60 and 90 s to quantify cell loss and activation. Results confirm preservation of ∼ 100% of monocytes, granulocytes and loss of ∼ 30% of lymphocytes (mostly CD3+/CD4+) with minimal activation. These results indicate feasibility of this approach for monocyte, granulocyte and lymphocyte (sub-populations) isolation based on size.







Similar content being viewed by others
References
S. E. Calvano et al., A network-based analysis of systemic inflammation in humans. Nature advanced online publication (2005)
J.P. Cobb et al., Application of genome-wide expression analysis to human health and disease. PNAS 102, 4801–4806 (2005)
R.J. Feezor et al., Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics 19, 247–254 (2004)
K. Laudanski et al., Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways. Proc Natl Acad Sci 103, 15564–15569 (2006). 10.1073/pnas.0607028103
T.W. Kuijpers et al., Membrane surface antigen expression on neutrophils: a reappraisal of the use of surface markers for neutrophil activation. Blood 78, 1105–1111 (1991)
J. Lundahl, G. Halldén, M. Hallgren, C.M. Sköld, J. Hed, Altered expression of CD11b/CD18 and CD62L on human monocytes after cell preparation procedures. J Immunol Meth 180, 93–100 (1995)
M.G. Macey, D.A. McCarthy, S. Vordermeier, A.C. Newland, K.A. Brown, Effects of cell purification methods on CD11b and -selectin expression as well as the adherence and activation of leucocytes. J Immunol Meth 181, 211–219 (1995)
A. Neurauter et al., Advances in Biochemical Engineering/Biotechnology, in Cell Separation, vol. 106, ed. by Kumar Ashok, Galaev Igor, Mattiasson Bo (Springer Berlin, Heidelberg, 2007), pp. 41–73
J. Sleasman, B. Leon, L. Aleixo, M. Rojas, M. Goodenow, Immunomagnetic selection of purified monocyte and lymphocyte populations from peripheral blood mononuclear cells following cryopreservation. Clin Diagn Lab Immunol 4, 653–658 (1997)
L.B. Ujam et al., Isolation of monocytes from human peripheral blood using immuno-affinity expanded-bed adsorption. Biotechnol Bioeng 83, 554–566 (2003)
J. Yoon, A. Terada, H. Kita, CD66b regulates adhesion and activation of human eosinophils. J Immunol 179, 8454–8462 (2007)
S.S. Kuntaegowdanahalli, A.A. Bhagat, G. Kumar, I. Papautsky, Inertial microfluidics for continuous particle separation in spiral microchannels. Lab Chip 9, 2973–2980 (2009)
D. Di Carlo, J.F. Edd, D. Irimia, R.G. Tompkins, M. Toner, Equilibrium separation and filtration of particles using differential inertial focusing. Anal Chem 80, 2204–2211 (2008). 10.1021/ac702283m
A. Russom, A. K. Gupta, S. Nagrath, D. Di Carlo, J. F. Edd, M. Toner, Differential inertial focusing of particles in curved low-aspect-ratio microchannels. New J Phys 11 (2009)
J.V. Green, M. Radisic, S.K. Murthy, Deterministic lateral displacement as a means to enrich large cells for tissue engineering. Anal Chem 81, 9178–9182 (2009). 10.1021/ac9018395
L.R. Huang, E.C. Cox, R.H. Austin, J.C. Sturm, Continuous particle separation through deterministic lateral displacement. Science 304, 987–990 (2004). 10.1126/science.1094567
S. Kapishnikov, V. Kantsler, V. Steinberg, Continuous particle size separation and size sorting using ultrasound in a microchannel. J Stat Mech 2006, 1742–5468 (2006)
X. Chen, D.F. Cui, C.C. Liu, H. Li, Microfluidic chip for blood cell separation and collection based on crossflow filtration. Sens Actuators, B 130, 216–221 (2008)
H. Ji et al., Silicon-based microfilters for whole blood cell separation. Biomed Microdevices 10, 251–257 (2008). 10.1007/s10544-007-9131-x
S. Murthy, P. Sethu, G. Vunjak-Novakovic, M. Toner, M. Radisic, Size-based microfluidic enrichment of neonatal rat cardiac cell populations. Biomed Microdevices 8, 231–237 (2006)
P. Sethu, A. Sin, M. Toner, Microfluidic diffusive filter for apheresis (leukapheresis). Lab Chip 6, 83–89 (2006a)
A. Russom et al., Differential inertial focusing of particles in curved low-aspect-ratio microchannels. New J Phys 11, 075025 (2009b)
P. Sethu et al., Microfluidic isolation of leukocytes from whole blood for phenotype and gene expression analysis. Anal Chem 78, 5453–5461 (2006b)
Acknowledgements
The authors would like to thank Tim Andrews, phlebotomist, Division of Pediatric Hematology/Oncology for help with obtaining blood samples and helpful discussions. This work was supported through a proof-of-concept grant (POCG) through the Office of Technology Transfer and the Vice President of Research at the University of Louisville and by the National Science Foundation under Grant No. 0814194.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Parichehreh, V., Estrada, R., Kumar, S.S. et al. Exploiting osmosis for blood cell sorting. Biomed Microdevices 13, 453–462 (2011). https://doi.org/10.1007/s10544-011-9513-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10544-011-9513-y