Abstract
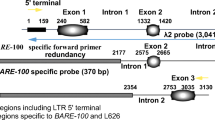
The JNK motif is repeated 4 000 times in an additional heterochromatin band. Flanking sequences of a JNK region, which form extra heterochromatin in 2R rye (Secale vavilovii Grossh.) chromosomes, were studied using a genome walking technique. The results clearly indicate that there were blocks of JNK sequences adjacent to the R173 family of repeated sequences. Moreover, it appears that the R173 are sequences flanking in both directions, i.e., upstream and downstream. Downstream, the R173 is adjacent to the JNKs in an anti-parallel orientation, whereas upstream it is adjacent in a parallel orientation. In order to confirm the presence of the R173 sequence, fluorescence in situ hybridization was carried out. Using both JNK and R173 molecular probes, overlapping hybridization signals in the 2RL pair of chromosomes were observed, indicating an identical location of the two sequence elements.
Similar content being viewed by others
Abbreviations
- DAPI:
-
4′,6-diamidino-2-phenylindole
- FISH:
-
fluorescence in situ hybridization
- SSC:
-
saline sodium citrate
- TBE:
-
Tris-borate-EDTA
References
Achrem, M., Kalinka, A., Rogalska, S.M.: Chromosomal localization of Ty1-copia i Ty3-gypsy retrotransposons in Secale vavilovii Grossh. using FISH. — In: Naganowska B., Kachlicki P., Krajewski P. (ed.): Genetics and Genomics for the Improvement of Crop Plants. Pp. 499–505. Institute of Plant Genetics, Polish Academy of Sciences, Poznan 2009.
Achrem, M., Rogalska, S.M., Kalinka, A.: Possible ancient origin of heterochromatic JNK sequences in chromosomes 2R of Secale vavilovii Grossh. — J. appl. Genet. 51: 1–8, 2010.
Chawla, R., DeMason, D.: Genome walking in pea: an approach to clone unknown flanking sequences. — Genetics 35: 3–5, 2003.
Cheng, Z.J., Murata, M.: A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. — Genetics 164: 665–672, 2003.
Cuadrado, A., Jouve, N.: Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. — J. Hered. 93: 339–345, 2002.
Eissenberg, J.C., Elgin, S.C.: The HP1 protein family: getting a grip on chromatin. — Curr. Opin. Genet. Dev. 10: 204–210, 2000.
Filippova, G.N.: Genetics and epigenetics of the multifunctional protein CTCF. — Curr. Topics dev. Biol. 80: 337–360, 2008.
Gniadkowski, M., Fiett, J., Borsuk, P., Hoffman-Zacharska, D., Stępień, P.P., Bartnik, E.: Structure and evolution of 5S rRNA genes and pseudogenes in the genus Aspergillus. — J. mol. Evol. 33: 175–178, 1991.
González-García, M., Cuacos, M., González-Sánchez, M., Puertas, M.J., Vega J.M.: Painting the rye genome with genome-specific sequences. — Genome 54: 555–564, 2011.
Grewal, S.C.R., Elgin, S.I.S.: Heterochromatin: new possibilities for inheritance of structure. — Curr. Opin. Genet. Dev. 12: 178–187, 2002.
Guidet, F., Rogowsky, P., Taylor, C., Song, W., Langridge, P.: Cloning and characterisation of a new rye specific repeated sequence. — Genome 34: 81–87, 1991.
Herold, M., Bartkuhn, M., Renkawitz, R.: CTCF: insights into insulator function during development. — Development 139: 1045–1057, 2012.
Huang, G., Zhang, L., Birch, R.G.: Rapid amplification and cloning of Tn5 flanking fragments by inverse PCR. — Lett. appl. Microbiol. 31: 149–153, 2000.
Kalendar, R., Lee, D., Schulman, A.H.: Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. — Genomics 98: 137–144, 2011.
Kellum, R., Schedl, P.: A position-effect assay for boundaries of higher order chromosomal domains. — Cell 64: 941–950, 1991.
Leoni, C., Gallerani, R., Ceci, L.R.: A genome walking strategy for the identification of eukaryotic nucleotide sequences adjacent to known regions. — BioTechniques 44: 229–235, 2008.
Levano-Garcia, J., Verjovski-Almeida, S., Da Silva, A.C.R.: Mapping transposon insertion sites by touchdown PCR and hybrid degenerate primers. — BioTechniques 38: 225–229, 2005.
Martins, C., Wasko, A.P., Oliveira, C., Porto-Foresti, F., Parise-Maltempi, P.P., Wright, J.M., Foresti, F.: Dynamics of 5S rDNA in the tilapia (Oreochromis niloticus) genome: repeat units, inverted sequences, pseudogenes and chromosome loci. — Cytogenet. Genome Res. 98: 78–85, 2002.
Martins, C, Wasko, A.P.: Organization and evolution of 5S ribosomal DNA in the fish genome. — In: Williams, C.R. (ed.): Focus on Genome Research. Pp. 335–363. Nova Science Publishers. Hauppauge — New York 2004.
Nagaki, K., Tsujimoto, H., Saskuma, T.: A novel repetitive sequence, termed the JNK repeat family, located on an extra heterochromatic region of chromosome 2R of Japanese rye. — Chromosome Res. 6: 95–101, 1999.
Raskina, O., Belyayev, A., Nevo, E.: Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. — Chromosome Res. 12: 153–161, 2004.
Rogalska, S.M., Achrem, M., Stróżycki, P.: The pScJNK1 repeated sequences identified on an extra heterochromatin band on chromosome 2R of Secale vavilovii Grossh. lines. — Biol. Bull. 38: 15–19, 2001.
Rogalska, S.M., Achrem, M., Słomińska-Walkowiak, R., Filip, E., Skuza, L., Pawłowska, J., Apolinarska, B.: Poly-morphism of heterochromatin bands on chromosomes of rye Secale vavilovii Grossh. lines. — Acta biol. cracow. Ser. bot. 44: 111–117, 2002a.
Rogalska, S.M., Apolinarska, B., Achrem, M., Słomińska-Walkowiak, R., Skuza, L., Filip, E.: Meiotic behavior of chromosomes in PMCs in plants of Secale vavilovii Grossh. lines with additional heterochromatin in chromosome 2R. — Acta biol. cracow. Ser. bot. 44: 119–124, 2002b.
Rogowsky, P., Manning, S., Liu, J.Y., Langidge, P.: The R173 family of rye-specific repetitive DNA sequences: a structural analysis. — Genome 34: 88–95, 1991.
Rogowsky, P.M., Liu, J.Y., Manning, S., Taylor, Ch., Langridge, P.: Structural heterogeneity in the R173 family of rye-specific repetitive DNA sequences. — Plant mol. Biol. 20: 95–102, 1992.
Rozen, S., Skaletsky, H.J.: Primer3 on the www for general users and for biologist programmers. — In: Krawetz, S., Misener, S. (ed.): Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa 2000.
Schindelhauer, D., Schwarz, T.: Evidence for a fast, intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous alpha-satellite DNA array. — Genome Res. 12: 1815–1826, 2002.
Scott, K.C., Merrett, S.L., Willard, H.F.: A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. — Curr. Biol. 16: 119–129, 2006.
Sharma, A., Wolfgruber, T.K., Presting, G.G.: Tandem repeats derived from centromeric retrotransposons. — BMC Genomics 14: 142, 2013.
Simms, T.A., Miller, E.C., Buisson, N.P., Jambunathan, N., Donze, D.: The Saccharomyces cerevisiae TRT2 tRNA Thr gene upstream of STE6 is a barrier to repression in MATα cells and exerts a potential tRNA position effect in MATa cells. — Nucl. Acids Res. 32: 5206–5213, 2004.
Skuza, L., Rogalska, S.: The change of the structure of atp1 mitochondrial gene are connected with presence of additional heterochromatin in 2R chromosomes in Secale vavilovii Grossh. lines. — In: Börner, A., Kobijlski, B. (ed.): The European Cereals Genetics Monographs. Pp. 198–201. Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung, Gatersleben 2012.
Sun, F.L., Elgin, S.C.: Putting boundaries on silence. — Cell 99: 459–462, 1999.
Tek, A.L., Song, J., Macas, J., Jiang, J.: Sobo, a recently amplified satellite repeat of potato, and its implications for the origin of tandemly repeated sequences. — Genetics 170: 1231–1238, 2005.
Terauchi, R., Kahl, G.: Rapid isolation of promoter sequences by TAIL-PCR: the 5′-flanking regions of Pal and Pgi genes from yams (Dioscorea). — Mol. gen. Genet. 263: 554–560, 2000.
Trindade, L.M., Horvath, B., Bachem, C., Jacobsen, E., Visser, R.G.F.: Isolation and functional characterization of stolon specific promoter from potato (Solanum tuberosum L.). — Gene 303: 77–87, 2003.
Van Ryk, D.I., Lee, Y., Nazar, R.N.: Efficient expression and utilization of a mutant 5S rRNA in Saccharomyces cerevisiae. — J. biol. Chem. 265: 8377–8381, 1990.
Xu, W., Shang, Y., Zhu, P., Zhai, Z., He, J., Huang, K., Luo Y.: Randomly broken fragment PCR with 5′ end-directed adaptor for genome walking. — Sci. Rep. 3: 3456, 2013.
Yan, Y., An, C., Li, L., Gu, J., Tan, G., Chen, Z.: T-linker-specific ligation PCR (T-linker PCR): an advanced PCR technique for chromosome walking or for isolation of tagged DNA ends. — Nucl. Acids Res. 31: e68, 2003.
Zhang, Z., Schwartz, S., Wagner, L., Miller, W.: A greedy algorithm for aligning DNA sequences. — J. Comput. Biol. 7: 203–214, 2000.
Zhao, X., Lu, J., Zhang, Z., Hu, J., Huang, S., Jin W.: Comparison of the distribution of the repetitive DNA sequences in three variants of Cucumis sativus reveals their phylogenetic relationships. — J. Genet. Genomics 38: 39–45, 2011.
Zlatanova, J., Caiafa, P.: CCCTC-binding factor: to loop or to bridge. — Cells Mol. Life Sci. 66: 1647–1660, 2009.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgments: This work was financially supported by the National Science Centre, Poland (Grant No. NN 310 436038).
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kalinka, A., Achrem, M. Analysis of the flanking sequences of the heterochromatic JNK region in Secale vavilovii chromosomes. Biol Plant 59, 637–644 (2015). https://doi.org/10.1007/s10535-015-0531-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-015-0531-0