Abstract
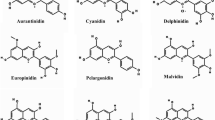
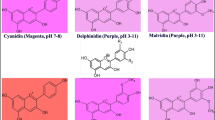
Chili pepper (Capsicum annuum L.) cv. Árbol and Uvilla fruits differing in anthocyanin contents were analyzed to characterize the accumulation patterns. The maximum accumulation of the aglycon delphinidin occurred 20 days postanthesis (DPA) with higher content in Uvilla than in Árbol fruits. Regarding the cDNA library, 9 186 cDNA clones were selected. The clones with high homology to genes concerning anthocyanin biosynthesis, such as encoding chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′,5′-hydroxylase (F3′5′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), UDP Glc-flavonoid 3-O-gluco-syl transferase (UFGT), and also those possibly involved in anthocyanin transport into the vacuoles, an anthocyanin permease (ANP) and a glutathione S-transferase (GST) were used for gene expression analysis. In general, the expression of all investigated genes was developmentally regulated in both Árbol and Uvilla. CHS and CHI transcripts were expressed at the maximal level at 10 DPA, and then consistently declined throughout fruit development. F3′5′H, DFR, UFGT and GST expression exhibited a positive correlation with anthocyanin accumulation, and the highest transcript levels were detected prior to or by the time of maximum anthocyanin accumulation, depending on the chili pepper type. Pericarp fruit tissues from cv. Tampiqueño 74, an anthocyanin non-accumulator, also showed CHS, CHI, F3H, ANS and ANP expression at some developmental stages.
Similar content being viewed by others
Abbreviations
- ANP:
-
anthocyanin permease
- ANS:
-
anthocyanidin synthase
- CHI:
-
chalcone isomerase
- CHS:
-
chalcone synthase
- DFR:
-
dihydroflavonol 4-reductase
- DPA:
-
days post anthesis
- F3H:
-
flavanone 3-hydroxylase
- F3′5′H:
-
flavonoid 3′,5′-hydroxylase
- Glc:
-
glucose
- GST:
-
glutathione S-transferase
- UDP:
-
uridindiphosphate
- UFGT:
-
UDP Glc-flavonoid 3-O-glucosyl transferase
References
Ahmed, N., Maekawa, M., Noda, K.: Anthocyanin accumulation and expression pattern of anthcyanin biosynthesis gene in developing wheat coleoptiles. — Biol Plant. 53: 223–228, 2009.
Ben Chaim, A., Borovsky, Y., De Jong, W., Paran, I.: Linkage of the A locus for the presence of anthocyanin and fs10.1 a major fruit-shape QTL in pepper. — Theor. appl. Genet. 106: 889–894, 2003.
Borovsky, Y., Oren-Shamir, M., Ovadia, R., De Jong, W., Paran, I.: The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. — Theor. appl. Genet. 109: 23–29, 2004.
Britsch, L., Dedio, J., Saedler, H., Forkmann, G.: Molecular characterization of flavanone 3β-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues. — Eur. J. Biochem. 217: 745–754, 1993.
Castellarin, S.D., Di Gaspero, G., Marconi, R., Nonis, A., Peterlunger, E., Paillard, S., Adam-Blondon, A.F., Testolin, R.: Colour variation in red grapevines (Vitis vinifera L.): genomic organisation, expression of flavonoid 3’- hydroxylase, flavonoid 3′,5′-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. — BMC Genomics 7: 1–12, 2006.
Conn, S., Curtin, C., Bézier, A., Franco, C., Zhang, W.: Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. — J. exp. Bot. 59: 3621–3634, 2008.
De Jong, W.S., Eannetta, N.T., De Jong, D.M., Bodis, M.: Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. — Theor. appl. Genet. 108: 423–432, 2004.
Deluc, L., Bogs, J., Walker, A.R., Ferrier, T., Decendit, A., Merillon, J.M., Robinson, S.P., Barrieu, F.: The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. — Plant Physiol. 147: 2041–2053, 2008.
De Vetten, N., Quattrocchio, F., Mol, J., Koes, R.: The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. — Genes Dev. 11: 1422–1434, 1997.
Espley, R.V., Hellens, R.P., Putterill, J., Stevenson, D.E., Kutty- Amma, S., Allan, A.C.: Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. — Plant J. 49: 414–427, 2007.
Griesser, M., Hoffmann, T., Bellido, M.L., Rosati, C., Fink, B., Kurtzer, R., Aharoni, A., Muñoz-Blanco, J., Schwab, W.: Redirection of flavonoid biosynthesis through the downregulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. — Plant Physiol. 146: 1528–1539, 2008.
Grotewold, E.: The genetics and biochemistry of floral pigments. — Annu. Rev. Plant Biol. 57: 761–780, 2006.
Holton, T.A., Cornish, E.: Genetics and biochemistry of anthocyanin biosynthesis. — Plant Cell 7: 1071–1083, 1995.
Jaakola, L., Määttä, K., Pirttilä, A.M., Törrönenm, R., Kärenlampi, S., Hohtola, A.: Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. — Plant Physiol. 130: 729–739, 2002.
Jez, J.M., Bowman, M.E., Dixon, R.A., Noel, J.P.: Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. — Nat. Struct. Biol. 7: 786–791, 2000.
Kitamura, S., Shikazono, N., Tanaka, A.: TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. — Plant J. 37: 104–114, 2004.
Lamy, S., Lafleur, R., Bédard, V., Moghrabi, A., Barrette, S., Gingras, D., Béliveau, R.: Anthocyanidins inhibit migration of glioblastoma cells: Structure-activity relationship and involvement of the plasminolytic system. — J. cell. Biochem. 100: 100–111, 2007.
Lightbourn, G.J., Griesbach, R.J., Novotny, J.A., Clevidence, B.A., Rao, D.D., Stommel, J.R.: Effects of anthocyanin and carotenoid combinations on foliage and immature fruit color of Capsicum annuum L. — J. Hered. 99: 105–111, 2008.
Lo Piero, A.R., Puglisi, I., Rapisarda, P., Petrone, G.: Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. — J. agr. Food Sci. 53: 9083–9088, 2005.
Marin, A., Ferreres, F., Tomás-Barberán, F.A., Gil, M.I.: Characterization and quantitation of antioxidant constituents of sweet pepper (Capsicum annuum L.). — J. agr. Food Chem. 52: 3861–3869, 2004.
Marrs, K.A., Alfenito, M.R., Lloyd, A.M., Walbot, V.: A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. — Nature 375: 397–400, 1995.
Mathews, H., Clendennen, S.K., Caldwell, C.G., Liu, X.L., Connors, K., Matheis, N., Schuster, D.K., Menasco, D.J., Wagoner, W., Lightner, J., Wagner, D.R.: Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. — Plant Cell 15: 1689–1703, 2003.
Morita, Y., Saitoh, M., Hoshino, A., Nitasaka, E., Iida, S.: Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. — Plant Cell Physiol. 7: 457–470, 2006.
Mueller, L.A., Goodman, C.D., Silady, R.A., Walbot, V.: AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. — Plant Physiol. 123: 1561–1570, 2000.
Nyman, N.A., Kumpulainen, J.T.: Determination of anthocyanidins in berries and red wine by high-performance liquid chromatography. — J. agr. Food Chem. 49: 4183–4187, 2001.
Preston, J., Wheeler, J., Heazlewood, J., Li, S.F., Parish, R.W.: AtMYB32 is required for normal pollen development in Arabidopsis thaliana. — Plant J. 40: 979–995, 2004.
Quattrocchio, F., Wing, J.F., Leppen, H., Mol, J., Koes, R.E.: Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. — Plant Cell 5: 1497–1512, 1993.
Reddy, M.K., Alexander-Lindo, R.L., Nair, M.G.: Relative inhibition of lipid peroxidation, cyclooxygenase enzymes, and human tumor cell proliferation by natural food colors. — J. agr. Food Chem. 53: 9268–9273, 2005.
Reif, H.J., Niesbach, U., Deumling, B., Saedler, H.: Cloning and analysis of two genes for chalcone synthase from Petunia hybrida. — Mol. gen. Genet. 199: 208–215, 1985.
Sadilova, E., Stintzing, F.C., Carle, R.: Anthocyanins, colour and antioxidant properties of eggplant (Solanum melongena L.) and violet pepper (Capsicum annuum L.) peel extracts. — Z. Naturforsch. C 61: 527–535, 2006.
Spelt, C., Quattrocchio, F., Mol, J.N., Koes, R.: Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. — Plant Cell 12: 1619–1632, 2000.
Stommel, J.R., Lightbourn, G.J., Winkel, B.S., Griesbach, R.J.: Transcription factor families regulate the anthocyanin biosynthetic pathway in Capsicum annuum. — J. amer. Soc. hort. Sci. 134: 244–251, 2009.
Wu, X., Prior, R.L.: Identification and characterization of anthocyanins by high-performance liquid chromatographyelectrospray ionization-tandem mass spectrometry in common foods in the United States: vegetables, nuts and grains. — J. agr. Food Chem. 53: 3101–3113, 1998.
Zhang, Q., Su, L.-J., Chen, J.-W., Zeng, X.-Q., Sun, B.-Y., Peng, C.-L.: The antioxidative role of anthocyanins in Arabidopsis under high-irradiance. — Biol. Plant. 56: 97–104, 2012.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgements: We acknowledge the National Council for Science and Technology (Conacyt; Mexico) for the fellowship to César Aza-González. The authors would like to thank Hamlet Avilés-Arnaut for his assistance to carry out the semi-quantitative expression analyses. This work was partially supported by grants from Fondos Mixtos Guanajuato (projects GTO-04-C02-24 and GTO-2005-C02-50), Conacyt (Project 55264) and SAGARPA (project 11806).
Rights and permissions
About this article
Cite this article
Aza-González, C., Herrera-Isidrón, L., Núñez-Palenius, H.G. et al. Anthocyanin accumulation and expression analysis of biosynthesis-related genes during chili pepper fruit development. Biol Plant 57, 49–55 (2013). https://doi.org/10.1007/s10535-012-0265-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-012-0265-1