Abstract
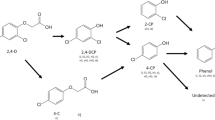
Metamifop is universally used in agriculture as a post-emergence aryloxyphenoxy propionate herbicide (AOPP), however its microbial degradation mechanism remains unclear. Consortium ME-1 isolated from AOPP-contaminated soil can degrade metamifop completely after 6 days and utilize it as the carbon source for bacterial growth. Meanwhile, consortium ME-1 possessed the ability to degrade metamifop stably under a wide range of pH (6.0–10.0) or temperature (20–42 °C). HPLC–MS analysis shows that N-(2-fluorophenyl)-2-(4-hydroxyphenoxy)-N-methyl propionamide, 2-(4-hydroxyphenoxy)-propionic acid, 6-chloro-2-benzoxazolinone and N-methyl-2-fluoroaniline, were detected and identified as four intermediate metabolites. Based on the metabolites identified, a putative metabolic pathway of metamifop was proposed for the first time. In addition, the consortium ME-1 was also able to transform or degrade other AOPP such as fenoxaprop-p-ethyl, clodinafop-propargyl, quizalofop-p-ethyl and cyhalofop-butyl. Moreover, the community structure of ME-1 with lower microbial diversity compared with the initial soil sample was investigated by high throughput sequencing. β-Proteobacteria and Sphingobacteria were the largest class with sequence percentages of 46.6% and 27.55% at the class level. In addition, 50 genera were classified in consortium ME-1, of which Methylobacillus, Sphingobacterium, Bordetella and Flavobacterium were the dominant genera with sequence percentages of 25.79, 25.61, 14.68 and 9.55%, respectively.






Similar content being viewed by others
References
Abraham J, Silambarasan S (2013) Biodegradation of chlorpyrifos and its hydrolyzing metabolite 3,5,6-trichloro-2-pyridinol by Sphingobacterium sp. JAS3. Process Biochem 48:1559–1564
Amareshwari P, Bhatia M, Venkatesh K, Rani AR, Ravi GV, Bhakt P, Nair AS (2015) Isolation and characterization of a novel chlorpyrifos degrading flavobacterium species EMBS0145 by 16S rRNA gene sequencing. Interdiscip Sci 7:1–6
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. doi:10.1093/bioinformatics/btu170
Burrows HD, Santaballa JA, Steenken S (2002) Reaction pathways and mechanisms of photodegradation of pesticides. J Photoch Photobio B 67:71–108
Cai S, Cai TM, Liu S, Yang Q, He J, Chen LW, Hu J (2014) Biodegradation of N-methylpyrrolidone by Paracoccus sp. NMD-4 and its degradation pathway. Int Biodeter Biodeg 93:70–77
Cai Z, Zhang W, Li S, Ma J, Wang J, Zhao X (2015) Microbial degradation mechanism and pathway of the novel insecticide paichongding by a newly isolated Sphingobacterium sp. P1-3 from soil. J Agr Food Chem 63:3823–3829
Caracciolo AB, Topp E, Grenni P (2015) Pharmaceuticals in the environment: biodegradation and effects on natural microbial communities: a review. J Pharmaceut Biomed 106:25–36
Dong WL, Jiang S, Shi KW, Wang F, Li SH, Zhou J, Huang F, Wang YC, Zheng YX, Hou Y, Huang Y, Cui ZL (2015a) Biodegradation of fenoxaprop-p-ethyl (FE) by Acinetobacter sp. strain DL-2 and cloning of FE hydrolase gene afeH. Bioresour Technol 186:114–121
Dong WL, Hou Y, Xi XD, Wang F, Li ZK, Ye XF, Huang Y, Cui ZL (2015b) Biodegradation of fenoxaprop-ethyl by an enriched consortium and its proposed metabolic pathway. Int Biodeter Biodeg 97:159–167
Dong WL, Chen QZ, Hou Y, Li SH, Zhuang K, Huang F, Zhou J, Li ZK, Wang J, Fu L, Zhang ZG, Wang F, Cui ZL (2015c) Metabolic pathway involved in 2-methyl-6- ethylaniline degradation by Sphingobium sp. strain MEA3-1 and cloning of the novel flavin-dependent monooxygenase system meaBA. Appl Environ Microb 8:8254–8264
Dong WL, Wang F, Huang F, Wang YC, Zhou J, Ye XF, Li ZK, Hou Y, Huang Y, Ma JF, Jiang M, Cui ZL (2016) Metabolic pathway involved in 6-chloro-2-benzoxazolinone degradation by Pigmentiphaga sp. strain DL-8 and identification of the novel metal-dependent hydrolase CbaA. Appl Environ Microb 82:4169–4179. doi:10.1128/AEM.00532-16
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Flint HJ, Scott KP, Duncan SH, Louis P, Forano E (2012) Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306
Gennari M, Vincenti M, Nègre M, Ambrosoli R (1995) Microbial metabolism of fenoxaprop-ethyl. Pestic Sci 44:299–303
Glenn AE, Bacon CW (2009) FDB2 encodes a member of the arylamine N-acetyltransferase family and is necessary for biotransformation of benzoxazolinones by Fusarium verticillioides. J Appl Microbial 107:657–671
Hao DC, Song SM, Mu J, Hu WL, Xiao PG (2016) Unearthing microbial diversity of Taxus rhizosphere via MiSeq high-throughput amplicon sequencing and isolate characterization. Sci Rep. doi:10.1038/srep22006
Hou Y, Tao J, Shen WJ, Liu J, Li JQ, Li YF, Cao H, Cui ZL (2011) Isolation of the fenoxaprop-ethyl (FE)-degrading bacterium Rhodococcus sp. T1, and cloning of FE hydrolase gene feh. FEMS Microbial Lett 323:196–203
Hou Y, Li SH, Dong WL, Yuan Y, Wang YC, Shen WJ, Cui ZL (2015) Community structure of a propanil-degrading consortium and the metabolic pathway of Microbacterium sp. strain T4-7. Int Biodeter Biodeg 105:80–89
Hu LB, Yang JD, Zhou W, Yin YF, Chen J, Shi ZQ (2009) Isolation of a Methylobacillus sp. that degrades microcystin toxins associated with cyanobacteria. New Biotechnol 26:205–211
Janaki P, Chinnusamy C (2012) Determination of metamifop residues in soil under direct-seeded rice. Toxico Enviro Chem 94:1043–1052
Kim TJ, Chang HS, Kim JS, Hwang IT, Hong KS, Kim DW, Chung BJ (2003) Metamifop: mechanism of herbicidal activity and selectivity in rice and barnyardgrass. In The BCPC International Congress: Crop Science and Technology, Volumes 1 and 2. In: Proceedings of an International Congress held at the SECC, Glasgow, Scotland, UK
Li JQ, Liu J, Shen WJ, Zhao XL, Hou Y, Cao H, Cui ZL (2010) Isolation and characterization of 3,5,6-trichloro-2-pyridinol-degrading Ralstonia sp. strain T6. Bioresour Technol 101:7479–7483
Lin J, Chen J, Wang Y, Cai X, Wei X, Qiao X (2008) More toxic and photoresistant products from photodegradation of fenoxaprop-p-ethyl. J Agr Food Chem 56:8226–8230
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Mohd-Kamil NAF, Hussain NH, Mizad MB, Abdul-Talib S (2014) Enhancing performance of Sphingobacterium spiritivorum in bioremediation phenanthrene contaminated sand. Remediat J 24:119–128
Moon JK, Keum YS, Hwang EC, Park BS, Chang HR, Li QX, Kim JH (2007) Hapten syntheses and antibody generation for a new herbicide, metamifop. J Agr Food Chem 55:5416–5422
Moon JK, Kim JH, Shibamoto T (2010) Photodegradation pathways and mechanisms of the herbicide metamifop in a water/acetonitrile solution. J Agr Food Chem 58:12357–12365
Nam IH, Kim Y, Cho D, Kim JG, Song H, Chon CM (2015) Effects of heavy metals on biodegradation of fluorene by a Sphingobacterium sp. strain (KM-02) isolated from polycyclic aromatic hydrocarbon-contaminated mine soil. Environ Eng Sci 32:891–898
Nie ZJ, Hang BJ, Cai S, Xie XT, He J, Li SP (2011) Degradation of cyhalofop-butyl (CyB) by Pseudomonas azotoformans strain QDZ-1 and cloning of a novel gene encoding CyB-hydrolyzing esterase. J Agr Food Chem 59:6040–6046
Oike H, Aoki-Yoshida A, Kimoto-Nira H, Yamagishi N, Tomita S, Sekiyama Y, Kobori M (2016) Dietary intake of heat-killed Lactococcus lactis H61 delays age-related hearing loss in C57BL/6 J mice. Sci Rep. doi:10.1038/srep23556
Pandey P, Pathak H, Dave S (2016) Microbial ecology of hydrocarbon degradation in the soil: a review. Res J Environ Toxicol 10:1
Patel V, Jain S, Madamwar D (2012) Naphthalene degradation by bacterial consortium (DV-AL) developed from Alang-Sosiya ship breaking yard, Gujarat, India. Bioresour Technol 107:122–130
Roy S, Singh SB (2005) Phototransformation of clodinafop-propargyl. J Environ Sci Heal B 40:525–534
Schloss PD, Westcott SL (2011) Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microb 77:3219–3226
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Boil Evol 24:1596–1599
Wang F, Grundmann S, Schmid M, Dörfler U, Roherer S, Munch JC, Schroll R (2007) Isolation and characterization of 1,2,4-trichlorobenzene mineralizing Bordetella sp. and its bioremediation potential in soil. Chemosphere 67:896–902
Wang X, Hou ZG, Guo G, Zhao XF, Wang XH, Li ZB (2014) Photolysis kinetics and mechanism of metamifop. Chinese Agr Sci Bull 1:059
Whitacre DM, Ware GW (2004) The pesticide book. Meister Media Worldwide, Willoughby
Xiong J, Liu Y, Lin X, Zhang H, Zeng J, Hou J, Chu H (2012) Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbial 14:2457–2466
Xu J, Yao G, Liu D, Liu M, Wang P, Zhou Z (2016) Environmental fate of chiral herbicide fenoxaprop-ethyl in water-sediment microcosms. Sci Rep. doi:10.1038/srep26797
Zhang YH, Xu D, Liu JQ, Zhao XH (2014) Enhanced degradation of five organophosphorus pesticides in skimmed milk by lactic acid bacteria and its potential relationship with phosphatase production. Food Chem 164:173–178
Zhu Y, Lin X, Zhao F, Shi X, Li H, Li Y, Zhou G (2015) Erratum: meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci Rep 5:16546
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 21390200 and 31560031), the Jiangsu Province Natural Science Foundation for Youths (No. BK20140940), China Postdoctoral Science Foundation (No. 2016M601787), and the Postdoctoral Foundation of Jiangsu Province (No. 1601043B), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, A Project Supported by Program for New Century Excellent Talents in University, Program for Changjiang Scholars and Innovative Research Team in University (No. 06-A-047).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Dong, W., Liu, K., Wang, F. et al. The metabolic pathway of metamifop degradation by consortium ME-1 and its bacterial community structure. Biodegradation 28, 181–194 (2017). https://doi.org/10.1007/s10532-017-9787-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-017-9787-8