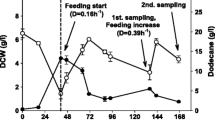
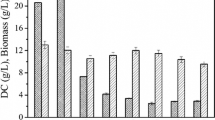
Abstract
Objectives
To explore Candida guilliermondii for the production of long-chain dicarboxylic acids (DCA), we performed metabolic pathway engineering aiming to prevent DCA consumption during β-oxidation, but also to increase its production via the ω-oxidation pathway.
Results
We identified the major β- and ω-oxidation pathway genes in C. guilliermondii and performed first steps in the strain improvement. A double pox disruption mutant was created that slowed growth with oleic acid but showed accelerated DCA degradation. Increase in DCA production was achieved by homologous overexpression of a plasmid borne cytochrome P450 monooxygenase gene.
Conclusion
C. guilliermondii is a promising biocatalyst for DCA production but further insight into its fatty acid metabolism is necessary.




Similar content being viewed by others
References
Craft DL, Madduri KM, Eshoo M, Wilson CR (2003) Identification and characterization of the CYP52 family of Candida tropicalis ATCC 20336, important fort he conversion of fatty acids and alkanes to alpha, omega-dicarboxylic acids. Appl Environ Microbiol 69:5983–5991
Defosse TA, Melin C, Obando Montoya EJ, Lanoue A, Foureau E et al (2014) A new series of vectors for constitutive, inducible or repressible gene expression in Candida guilliermondii. J Biotechnol 180:37–42
Foureau E, Courdavault V, Rojas LF, Dutilleul C, Simkin AJ et al (2013) Efficient gene targeting in a Candida guilliermondii non-homologous end-joining pathway-deficient strain. Biotechnol Lett 35:1035–1043
Gabriel F, Accoceberry I, Bessoule J-J, Salin B, Lucas-Guérin M (2014) A Fox2-dependent fatty acid β-oxidation pathway coexists both in peroxisomes and mitochondria of the ascomycete yeast Candida lusitaniae. PLoS ONE. doi:10.1371/journal.pone.0114531
Hara A, Ueda T, Matsui T, Arie M, Saeki H et al (2001) Repression of fatty-acyl-CoA oxidase-encoding gene expression is not necessarily a determinant of high-level production of dicarboxylic acids in industrial dicarboxylic-acid-producing Candida tropicalis. Appl Microbiol Biotechnol 56:478–485
Huf S, Krügener S, Hirth T, Rupp S, Zibek S (2011) Biotechnological synthesis of long-chain dicarboxylic acids as building blocks for polymers. Eur J Lipid Sci Technol 113:548–561
Masuda Y, Park SM, Ohta A, Takagi M (1995) Cloning and characterization of the POX2 gene in Candida maltosa. Gene 167:157–161
Millerioux Y, Clastre M, Simkin AJ, Courdavault V, Marais E (2011) Drug-resistant cassettes for efficient transformation of Candida guilliermondii wild-type strains. FEMS Yeast Res 11:457–463
Papon N, Courdavault V, Clastre M (2014) Biotechnological potential of the fungal CTG clade species in the synthetic biology era. Trends Biotechnol 32:167–168
Picataggio S, Deanda K, Mielenz J (1991) Determination of Candida tropicalis acyl coenzyme A oxidase isozyme function by sequential gene disruption. Mol Cell Biol 9:4333–4339
Picataggio S, Rohrer T, Deanda K, Lanning D, Reynolds R, Mielenz J, Eirich LD (1992) Metabolic engineering of Candida tropicalis for the production of long-chain dicarboxylic acids. Nat Biotechnol 10:894–898
Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK (2006) Peroxisomal β-oxidation—a metabolic pathway with multiple functions. Biochim Biophys Acta 12:1412–1426
Sibirny AA, Boretsky YB (2009) Pichia guilliermondii. In: Satyanarayana T, Kunze G (eds) Yeast biotechnology: diversity and applications, 1st edn. Springer, Berlin, pp 1113–1134
Syed K, Mashele SS (2014) Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE. doi:10.1371/journal.pone.0095616
Wang HJ, Le Dall M-T, Waché Y, Laroche C, Belin J-M, Gaillardin C, Nicaud J-M (1999) Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol 181:5140–5148
Acknowledgements
We are thankful for the funding support by the 7th Framework Programme of the European Union within the joint project “BioConSept” (Grant Agreement Number: 289194).
Supporting information
Supplementary Fig. 1—Overview ω-oxidation and β-oxidation pathway in yeasts.
Supplementary Fig. 2—Construction of pox1 gene disruption cassette.
Supplementary Fig. 3—Construction of pox1 and pox2 gene disruption cassette.
Supplementary Fig. 4—POX3 gene expression study.
Supplementary Fig. 5—Bioconversion of oleic acid by C. guilliermondii mutants overexpressing ω-oxidation pathway genes.
Supplementary Table 1—List of abbreviations and accession numbers of genes.
Supplementary Table 2—List of primers used for this study.
Supplementary Table 3—Percentage of sequence similarities between Pox homologous.
Supplementary Table 4—Percentage of sequence similarities between Cyp52 homologous.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 (EPS 123 kb)
Overview ω-oxidation and β-oxidation pathway in yeasts. Fatty acids are converted to the corresponding dicarboxylic acid via the microsomal ω-oxidation pathway and subsequently degraded through the peroxisomal β-oxidation. Acyl-CoA is further metabolized in the tricarboxylic and glyoxylate cycle. Involved enzymes and their known abbreviations are shown
Supplementary Fig. 2 (EPS 142 kb)
Construction of pox1 gene disruption cassette. a The sat1-resistance cassette was cloned into the AflII and SnaBI site of the plasmid pJet-pox1-ko containing a 7.1 kb C. guilliermondii genomic fragment including the POX1 gene and a 2.5 kb 3’ and 5’ POX1 sequence. b The 5.3 kb disruption cassette was released by digestion with PvuII and SpeI containing 1.5 and 1.9 kb homologous 5′ and 3′ flanking regions for targeted integration in the C. guilliermondii genome. c Correct integration was verified by PCR from isolated genomic DNA using the primer pair KOpox1_F2 (located in the genomic DNA, upstream and outside of the integrated deletion cassette) and KOpox1_R2 (located within the sat-1 resistance cassette) resulting in an 2.7 kb long amplicon, and KOpox1_F3 (located in the sat-1 resistance cassette) and KOpox1_R3 (located in the genomic DNA, downstream and outside the integrated deletion cassette) amplificating a 2.5 kb fragment in the correct pox1 disruption mutant
Supplementary Fig. 3 (EPS 166 kb)
Construction of pox1 and pox2 gene disruption cassette. a The sat1-resistance cassette was cloned into the AatII sites of the plasmid pJet-pox1,2-ko containing a 8.2 kb C. guilliermondii genomic fragment including the POX1 and POX2 gene and a 1.7 kb 3′ and 1.1 kb 5′ flanking region. b The 7.5 kb disruption cassette was released by digestion with BsRGI and EcoRV containing 2.7 and 1.8 kb homologous 5′ and 3′ flanking regions for targeted integration in the C. guilliermondii genome. c Correct integration was verified by PCR from isolated genomic DNA using the primer pair KOpox12_F2 (located in the genomic DNA, upstream and outside of the integrated deletion cassette) and KOpox1_R2 (located within the sat-1 resistance cassette) resulting in an 3.6 kb long amplicon, and KOpox1_F3 (located in the sat-1 resistance cassette) and KOpox12_R3 (located in the genomic DNA, downstream and outside the integrated deletion cassette) amplificating a 3.6 kb fragment in the correct pox1, pox2 double disruption mutant
Supplementary Fig. 4 (EPS 76 kb)
POX3 gene expression study. Fold change of POX3 gene expression in the Δpox1Δpox2 double mutant 5h after induction with oleic acid compared to expression in the parental strain KU141F1. Relative Expression levels were calculated using the Cgu actin1 gene as housekeeping gene and then converted to fold change values relative to their expression levels in KU141F1. Data are representative to three biological and three technical replicates. Calculated t test value 0.09. (Error bars represent SD)
Supplementary Fig. 5 (TIFF 8931 kb)
Bioconversion of oleic acid by C. guilliermondii mutants overexpressing ω-oxidation pathway genes. Thin layer chromatography showing fatty and dicarboxylic acid extractions from a bioconversion time course-experiment. The cells were cultivated for 96 h with oleic acid, which is converted into 1,18-octadecanedioic acid. a Oleic acid (OA) standard and 1,18-octadecanedioic acid standard (DCA). The DCA showed a slight reddish coloration after the staining. b DCA production with two independent CYP52A12 overexpressing mutants (cyp12_3 and cyp12_1), together with two independent control strains (parental strain KU141F1 bearing the empty vector construct (EVC)). Differences in the DCA concentration between EVC and cyp12_over after 72 h are highlighted with asterisks. c, d showing exemplary one of the biological duplicates of the CYP52A15 and NCP overexpressing mutants, respectively
Rights and permissions
About this article
Cite this article
Werner, N., Dreyer, M., Wagner, W. et al. Candida guilliermondii as a potential biocatalyst for the production of long-chain α,ω-dicarboxylic acids. Biotechnol Lett 39, 429–438 (2017). https://doi.org/10.1007/s10529-016-2264-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2264-3