Abstract
Objectives
To investigate the use of genome shuffling to generate recombinants from previously generated hydrolysates-tolerant strains to improve tolerance of Saccharomyces cerevisiae to one or more inhibitory by-products present in lignocellulosic hydrolysates.
Results
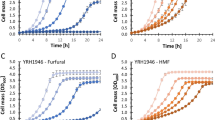
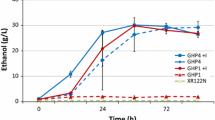
Recombinants of previously evolved strains of S. cerevisiae were generated and analyzed for their relative performance in the individual inhibitors furfural, acetic acid, 5-(hydroxymethyl)-furfural (HMF) and in synthetic hydrolysates. One recombinant exhibited a 100 % fitness increase in the presence of HMF as compared to the wild-type diploid, while another stain exhibited a 13 % fitness increase in the presence of furfural. Furthermore, for one of these recombinants, these increases in fitness were specific to the inhibitor HMF and to synthetic hydrolysates rather than being due to a general increase in fitness. Mutations present in the evolved hydrolysates-tolerant mutants were identified via whole-genome resequencing.
Conclusion
Recombinants of S. cerevisiae were produced with increased tolerance to inhibitory by-products present in hydrolysates of lignocellulosic biomass and identified potential genetic determinants associated with this phenotype.


Similar content being viewed by others
References
Almario MP, Reyes LH, Kao KC (2013) Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol Bioeng 110:2616–2623
Chen XZ, Peng JB, Cohen A, Nelson H, Nelson N, Hediger MA (1999) Yeast SMF1 mediates H(+)-coupled iron uptake with concomitant uncoupled cation currents. J Biol Chem 274:35089–35094
Ge J, Zhao J, Zhang L, Zhang M, Ping W (2014) Construction and analysis of high-ethanol-producing fusants with co-fermentation ability through protoplast fusion and double labeling technology. PLoS One 9:e108311
Heer D, Sauer U (2008) Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microbial Biotechnol 1:497–506
Kao KC, Sherlock G (2008) Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet 40:1499–1504
Lawford HG, Rousseau JD (1998) Improving fermentation performance of recombinant Zymomonas in acetic acid-containing media. Appl Biochem Biotechnol 70–72:161–172
Li Q, Song J, Peng S, Wang JP, Qu GZ, Sederoff RR, Chiang VL (2014) Plant biotechnology for lignocellulosic biofuel production. Plant Biotechnol J 12:1174–1192
Mussatto SI, Roberto IC (2004) Alternatives for detoxification of diluted-acid lignocellulosic hydrolyzates for use in fermentative processes: a review. Bioresour Technol 93:1–10
Palmqvist E, Hahn-Hägerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 74:25–33
Pasha C, Kuhad RC, Rao LV (2007) Strain improvement of thermotolerant Saccharomyces cerevisiae VS strain for better utilization of lignocellulosic substrates. J Appl Microbiol 103:1480–1489
Reyes LH, Almario MP, Winkler J, Orozco MM, Kao KC (2012) Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metabol Eng 14:579–590
Schnettler R, Zimmermann U, Emeis CC (1984) Large-scale production of yeast hybrids by electrofusion. FEMS Microbiol Lett 24:81–85
Shui ZX, Qin H, Wu B, Tan FR, Wang JL, Tang XY, Dai LC, Hu GQ, He MX, Ruan ZY, Wang LS (2015) Adaptive laboratory evolution of ethanologenic Zymomonas mobilis strain tolerant to furfural and acetic acid inhibitors. Appl Microbiol Biotechnol 99(13):5739–5748
Singh R, Shukla A, Tiwari S, Srivastava M (2014) A review on delignification of lignocellulosic biomass for enhancement of ethanol production potential. Renew Sustain Energy Rev 32:713–728
Supek F, Supekova L, Nelson H, Nelson N (1996) A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA 93:5105–5110
Taylor MP, Mulako I, Tuffin M, Cowan D (2012) Understanding physiological responses to pre-treatment inhibitors in ethanologenic fermentations. Biotechnol J 7:1169–1181
van der Pol EC, Bakker RR, Baets P, Eggink G (2014) By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for (bio)chemicals and fuels. Appl Microbiol Biotechnol 98:9579–9593
Xiao H, Zhao H (2014) Genome-wide RNAi screen reveals the E3 SUMO-protein ligase gene SIZ1 as a novel determinant of furfural tolerance in Saccharomyces cerevisiae. Biotechnol Biofuel 7:78
Xiros C, Olsson L (2014) Comparison of strategies to overcome the inhibitory effects in high-gravity fermentation of lignocellulosic hydrolysates. Biomass Bioenergy 65:79–90
Zha Y, Westerhuis JA, Muilwijk B, Overkamp KM, Nijmeijer BM, Coulier L, Smilde AK, Punt PJ (2014) Identifying inhibitory compounds in lignocellulosic biomass hydrolysates using an exometabolomics approach. BMC Biotechnol 14:22
Acknowledgments
This research was funded in part by NSF MCB-1054276.
Supporting Information
Supplementary Table 1—Primer Sequences.
Supplementary Fig. 1—Primers used for verification of SNPs identified from Next-Gen Sequencing and identification of SNPs in the diploid recombinants.
Supplementary Fig. 2—An example of growth data used to calculate maximum specific growth rates.
Supplementary Fig. 3—Growth rate of wild-type GFP-RFP in YNB media compiled from two independent experiments with three replicates each.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cheng, C., Almario, M.P. & Kao, K.C. Genome shuffling to generate recombinant yeasts for tolerance to inhibitors present in lignocellulosic hydrolysates. Biotechnol Lett 37, 2193–2200 (2015). https://doi.org/10.1007/s10529-015-1895-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1895-0