Abstract
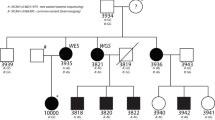
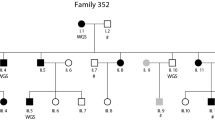
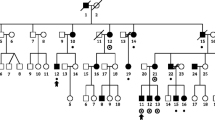
Dyslexia is a frequent neurodevelopmental learning disorder. To date, nine susceptibility loci have been identified, one of them being DYX9, located in Xq27. We performed the first French SNP linkage study followed by candidate gene investigation in dyslexia by studying 12 multiplex families (58 subjects) with at least two children affected, according to categorical restrictive criteria for phenotype definition. Significant results emerged on Xq27.3 within DYX9. The maximum multipoint LOD score reached 3,884 between rs12558359 and rs454992. Within this region, seven candidate genes were investigated for mutations in exonic sequences (CXORF1, CXORF51, SLITRK2, FMR1, FMR2, ASFMR1, FMR1NB), all having a role during brain development. We further looked for 5′UTR trinucleotide repeats in FMR1 and FMR2 genes. No mutation or polymorphism co-segregating with dyslexia was found. This finding in French families with Dyslexia showed significant linkage on Xq27.3 enclosing FRAXA, and consequently confirmed the DYX9 region as a robust susceptibility locus. We reduced the previously described interval from 6.8 (DXS1227–DXS8091) to 4 Mb also disclosing a higher LOD score.



Similar content being viewed by others
References
Abecasis GR, Wigginton JE (2005) Handling marker–marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet 77:754–767
Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Rapid-analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101
Aruga J, Mikoshiba K (2003) Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci 24:117–129
Bates T, Luciano M, Castles A, Coltheart M, Wright M, Martin N (2007) Replication of reported linkages for dyslexia and spelling and suggestive evidence for novel regions on chromosomes 4 and 17. Eur J Hum Genet 15:194–203
Brown WE, Eliez S, Menon V, Rumsey JM, White CD, Reiss AL (2001) Preliminary evidence of widespread morphological variations of the brain in dyslexia. Neurology 56:781–783
Cavalli-Sforza LL, King MC (1986) Detecting linkage for genetically heterogeneous diseases and detecting heterogeneity with linkage data. Am J Hum Genet 38:599–616
Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P et al (2005) Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet 76:59–581
Cornish KM, Kogan CS, Li L, Turk J, Jacquemont S, Hagerman RJ (2009) Lifespan changes in working memory in fragile X permutation males. Brain Cogn 69:551–558
Cottingham RW Jr., Idury RM, Schaffer AA (1993) Faster sequential genetic computations. Am J Hum Genet 53:252–263 A version of 4.1P Fastlink
Cunningham CL, Martinez Cerdeno V, Navarro E, Prakash A, Angelastro JM, Willemsen R et al (2011) Premutation CGG-repeat expansion of the Fmr1 gene impairs mouse neocortical development. Hum Mol Genet 20:64–79
Currier TA, Etchegaray MA, Haight JL, Galaburda AM, Rosen GD (2011) The effect of embryonic knockdown of the candidate dyslexia susceptibility gene DYX1C1 on the distribution of GABAergic neurons in the cerebral cortex. Neuroscience 172:535–546
Davis S, Weeks DE (1997) Comparison of nonparametric statistics for detection of linkage in nuclear families: single-marker evaluation. Am J Hum Genet 61:1431–1444
de Kovel GG, Hol FA, Heister JG, Willemen JJ, Sandkuijl LA, Franke B, Padberg GW (2004) Genomewide scan identifies susceptibility locus for dyslexia on Xq27 in an extended Dutch family. J Med Genet 41:652–657
DeFries JC, Alarcon M (1996) Genetic of specific reading disability. Ment Retard Dev Disabil Res Rev 2:39–47
Eliez S, Rumsey JM, Giedd JN, Schmitt JE, Patwardhan AJ, Reiss AL (2000) Morphological alteration of temporal lobe gray matter in dyslexia: an MRI study. J Child Psychol Psychiatry 41:637–644
ENCODE project consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74
Feingold E, Song KK, Weeks DE (2000) Comparison of allele-sharing statistics for general pedigrees. Genet Epidemiol Suppl 1:92–98
Fisher SE, Francks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR et al (2002) Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet 30:86–91
Flannery K, Liederman J, Daly L, Schultz J (2000) Male prevalence for reading disability is found in a large sample of black and white children free for ascertainment bias. J Int Neuropsychol Soc 6(4):433–444
Francks C, Paracchini S, Smith S, Richardson A, Scerri T, Cardon L et al (2004) A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet 75:1046–1058
Galaburda AM, Sherman GF, Rosen GD, Aboitiz F, Geschwind N (1985) Four consecutive patients with cortical anomalies. Ann Neurol 18:222–233
Galaburda AM, Menard M, Rosen GD (1994) Evidence for aberrant auditory anatomy in developmental dyslexia. Proc Natl Acad Sci USA 91:80110–80113
Habib M (2000) The neurological basis of developmental dyslexia: an overview and working hypothesis. Brain 123:2373–2399
Hallgren B (1950) Specific dyslexia (congenital word-blindness); a clinical and genetic study. Acta Psychiatr Neurol Suppl 65:1–287
Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmo J, Kaariainen H, Kere J (2005) The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet 1:467–474
Klingberg T, Hedehus M, Temple E, Salz T, Gabrieli JD, Moseley ME et al (2000) Microstructure of temporo-parietal white matter as a basis for reading ability: evidence from diffusion tensor magnetic resonance imaging. Neuron 25:493–500
Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363
Ladd P, Smith L, Rabaia N, Moore J, Georges S, Hansen S et al (2007) An antisense transcript spanning the CGG repeat region of FMR1 is up regulated in permutation carriers but silenced in full mutation individuals. Hum Mol Genet 16:3174–3187
Lyon G, Shaywitt SE, Shaywitz BA (2003) A definition of dyslexia. Ann Dyslexia 53:1–14
Massinen S, Hokkanen M-E, Matsson H, Tammimies K, Tapia-Páez I et al (2011) Increased expression of the dyslexia candidate gene DCDC2 affects length and signaling of primary cilia in neurons. PLoS One 6:e20580
Meng HY, Smith SD, Hager K, Held M, Liu J, Olson RK et al (2005) DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA 102:17053–17058
Muller-Myohsok B, Grimm T (1999) Linkage analysis and genetic models in dyslexia—considerations pertaining to discrete trait analysis and quantitative trait analyses. Eur Child Adolesc Psychiatry 8(Suppl 3):40–42
Pal DK, Greenberg BA (2002) Evaluating genetic heterogeneity in complex disorders. Hum Hered 53:216–226
Patterson N, Pricie AL, Reich D (2006) Population structure and gene analysis. PLoS Genet 2:2190
Pechansky VJ, Burbridge TJ, Volz AJ, Fiodella C, Wissner-Gross Z, Galaburda AM et al (2010) The effect of variation in expression of the candidate dyslexia susceptibility gene homolog Kiaa0319 on neuronal migration and dendritic morphology in the rat. Cereb Cortex 20:884–897
Poelmans G, Buitelaar J, Pauls D, Francke B (2011) A theoretical network for dyslexia: integrating available genetic findings. Mol Psychiatry 16:365–382
Price AL, Patterson NJ, Plenge RM et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 3:904–909
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 81:559–575
Raskind WH (2001) Current understanding of the genetic basis of reading and spelling disability. Learn Disabil Q 21:141–157
Rutter M, Caspi A, Fergussin D, Horwood LJ, Goodman R, Maughan B et al (2004) Sex differences in developmental reading disability: new findings from 4 epidemiological studies. JAMA 291:2007–2012
Scerri T, Schulte-Körne G (2010) Genetics of developmental dyslexia. Eur Child Adolesc Psychiatry 19:179–197
Schumacher J, Anthoni H, Dahdouh F, König IR, Hillmer AM, Kluck N et al (2006) Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet 78:52–62
Shaywitz SE (1998) Current concepts: dyslexia. N Engl J Med 338:307–312
Shaywitz BA, Shaywitz SE, Pugh KR, Mencl WE, Fullbright RK, Skudlarski P et al (2002) Disruption of posterior brain systems for reading in children with developmental dyslexia. Biol Psychiatry 52:101–110
Shugart YY, Chen L, Li R, Beaty T (2007) Family-based linkage disequilibrium tests using general pedigrees. Methods Mol Biol 376:141–149
Taipale M, Kaminen M, Nopola-Hemii J, Haltia T, Myllyluoma B, Lyytinen H et al (2003) A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci USA 100:11553–11558
R Development Core Team (2012) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria, ISBN 3-900051-07-0. http://www.R-project.org. Accessed 9 Feb 2012
Terwilliger JD, Goring HH (2000) Gene mapping in the 20th and 21st centuries: statistical methods data analysis and experimental design. Hum Biol 72(1):63–132
Vandermosten M, Boets B, Poelmans H, Sunaert S, Wouters J, Ghesquière P (2012) A tractography study in dyslexia: neuroanatomic correlates of orthographic, phonological and speech processing. Brain 135:935–948
Wigginton JE, Abecasis GR (2005) PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics 21:3445–3447
Ziegler A, König I, Deimel W, Plume E, Nöthen M, Propping P et al (2005) Developmental dyslexia—recurrence risk estimates from a German bi-center study using the single proband sib pair design. Hum Hered 59:136–143
Acknowledgments
This work was supported by a grant of the French Ministry of Education and Research. We thank D Raine, AS Galloux, AG Piller, G Turlotte, E Schweitzer, S Heath, and C Blanc-Tailleur for their valuable contribution. We are also very grateful to the families for their participation to the study.
Conflict of interest
All authors report no biomedical financial disclosures or potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Stacey Cherny.
M. Huc-Chabrolle and C. Charon contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huc-Chabrolle, M., Charon, C., Guilmatre, A. et al. Xq27 FRAXA Locus is a Strong Candidate for Dyslexia: Evidence from a Genome-Wide Scan in French Families. Behav Genet 43, 132–140 (2013). https://doi.org/10.1007/s10519-012-9575-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-012-9575-5